Our lab investigates humoral immunity and antibodies at the single-cell and molecular levels, with applications to infectious diseases, including SARS-CoV-2, influenza, HIV, and other challenging pathogens. We employ high-throughput single-cell sequencing and deep mutational scanning, computational modeling, and experimental virology to advance understanding and interventions.
Viral Evolution
Humoral Immune Response
Antibody Therapeutics
Our work maps SARS-CoV-2 mutational landscapes, and identifies evasion hotspots to anticipate future variants. We developed a high-throughput deep mutational scanning (DMS) technology, enabling large-scale mapping of antibody escaping mutation profiles, and developed a SARS-CoV-2 evolution prediction model was constructed based on population immune pressure calculated by DMS profiles from a large collection of neutralizing antibodies, allowing precise prediction of viral evolution. Achievements include developing models for RBD evolution, conducting deep mutational scanning, and characterizing emerging variants for their infectivity, antibody evasion, and antigenic properties.
- Mapped the strong immune escape mutations of Omicron BA.1 using the featured high-throughput DMS platform with structural and functional analyses integrated(Cao et al., Nature 2022).
- Analyzed antibody evasion by BA.2.12.1, BA.4, and BA.5, predicting dominance of these Omicron sublineages(Cao et al., Nature 2022).
- Predicted Omicron RBD evolution hotspots via high-throughput DMS and discovered the intrinsic relationship between immune imprinting and viral convergent evolution(Cao et al., Nature 2023).
- Continuous surveillance and characterization of emerging variants:
- BA.2.75:Cao et al., Cell Host & Microbe 2022
- BA.4.6/BF.7:Jian et al., Lancet Infectious Diseases 2022
- XBB.1.5:Yue et al., Lancet Infectious Diseases 2023
- "FLip" (EG.5 and HK.3):Jian et al. PLOS Pathogens 2023
- BA.2.86:Yang et al., Lancet Infectious Diseases 2023
- BA.2.87.1:Yang et al. Emerging Microbes & Infections 2024
- JN.1:Yang et al., Lancet Infectious Diseases 2024
- KP.3:Feng et al. Cell Discovery 2024
- KP.3.1.1 and XEC:Liu et al. Lancet Infectious Diseases 2025
- LF.7.2.1, NP.1 and LP.8.1:Liu et al., Lancet Infectious Diseases 2025
- BA.3.2, NB.1.8.1 and XFG:Guo et al., Lancet Infectious Diseases 2025
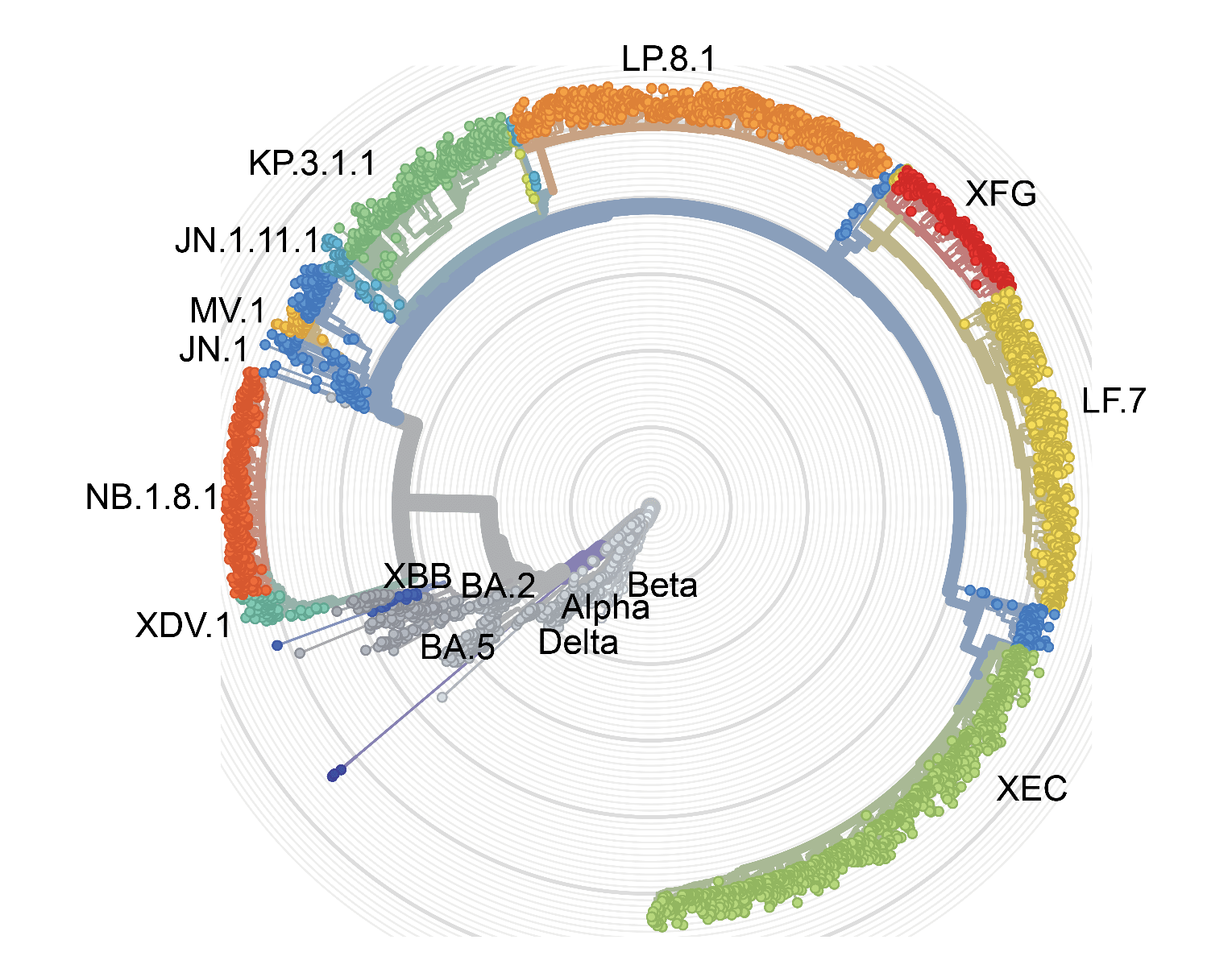
SARS-CoV-2 evolution and variants (Modified from nextstrain.org)
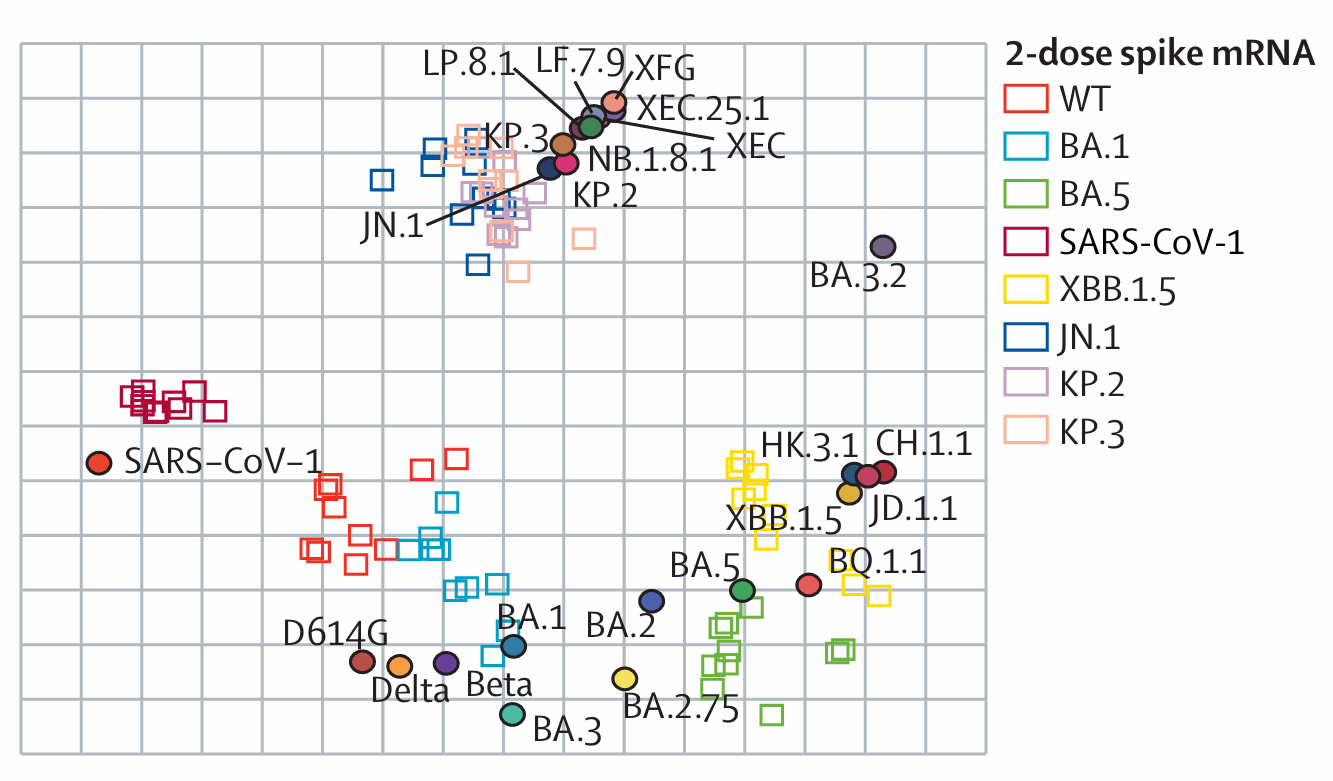
Antigenic map of SARS-CoV-2 major variants from mouse immunization

