[bioRxiv] Repeated Omicron infection alleviates SARS-CoV-2 immune imprinting
This is a copy of our previous thread on Twitter.
In our latest research released on bioRxiv, we found repeated Omicron infection/boosting alleviates WT vaccine-induced immune imprinting by generating many potent XBB-neutralizing Omicron-specific antibodies that target new RBD epitopes.
First, let's revisit the major concept of SARS-CoV-2 immune imprinting: When we experience a variant-vaccine boosting or breakthrough infection, our immune system will mainly recall WT vaccination-induced memory B cells and rarely produces variant-specific antibodies.
The problem caused by this concept is that when the boosting/infecting variant has a long antigenic distance to WT, the majority of memory B cells recalled will be those that target conserved and non-neutralizing epitopes, which will greatly hinder the antibody response.
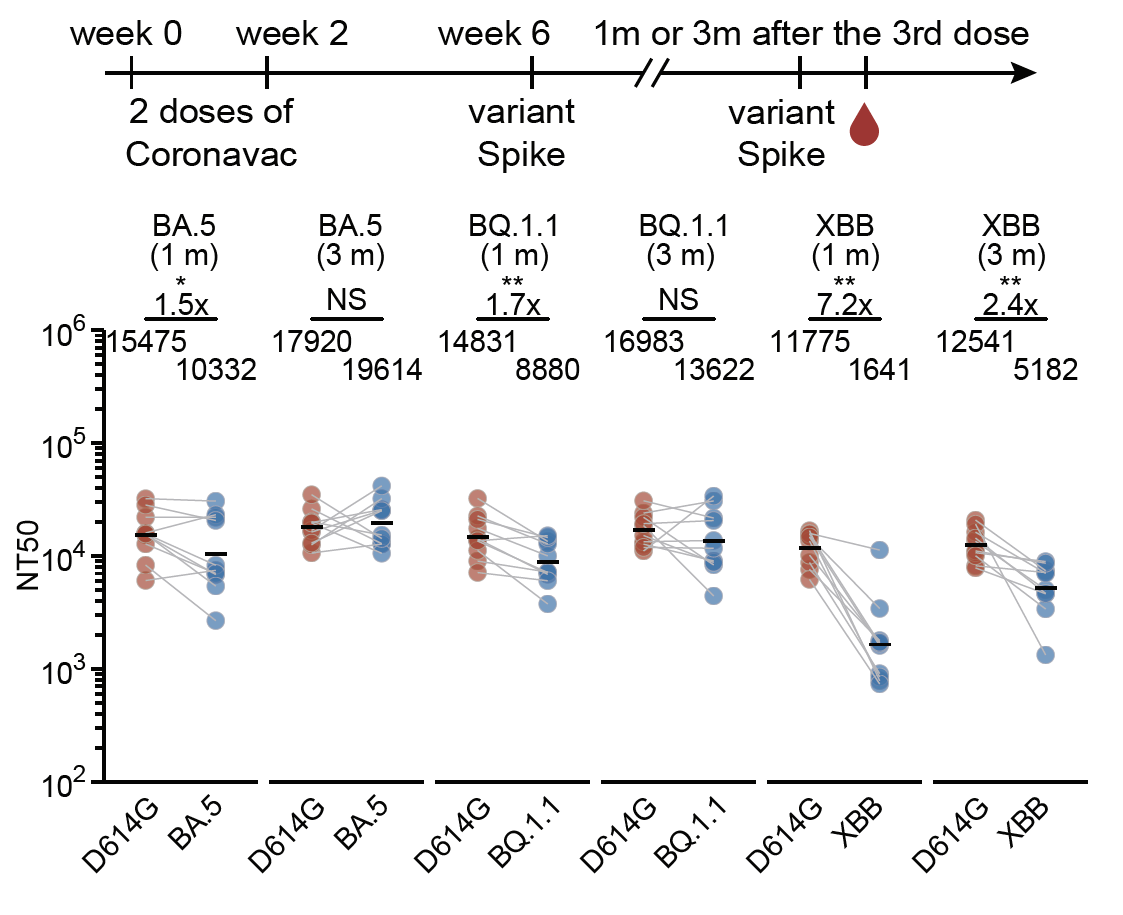
As a result, when the antigenic distance between the stimulating variant and WT increases (BA.1->BA.5->XBB), the overall serum neutralization response will substantially decrease, as shown in our data from WT-primed mice models and human cohorts.
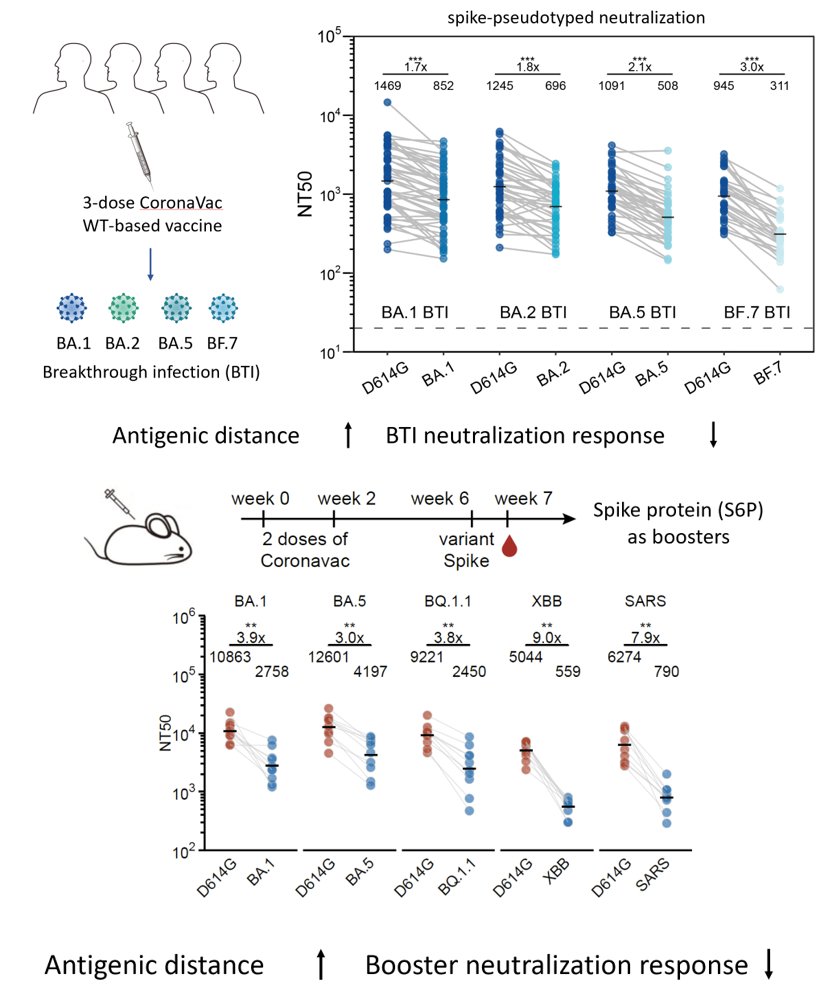
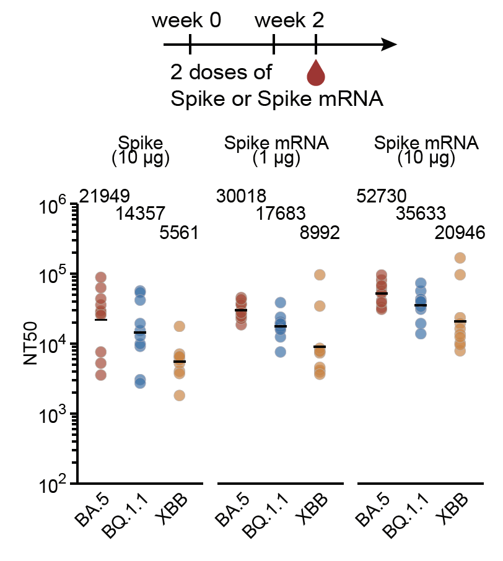
Increasing the interval between the WT priming and Omicron boosters or using mRNA boosters (higher immunogenicity) didn't help much to alleviate immune imprinting either unless you use a 10μg mRNA booster; however, that relatively high dosage is impractical in the real world.
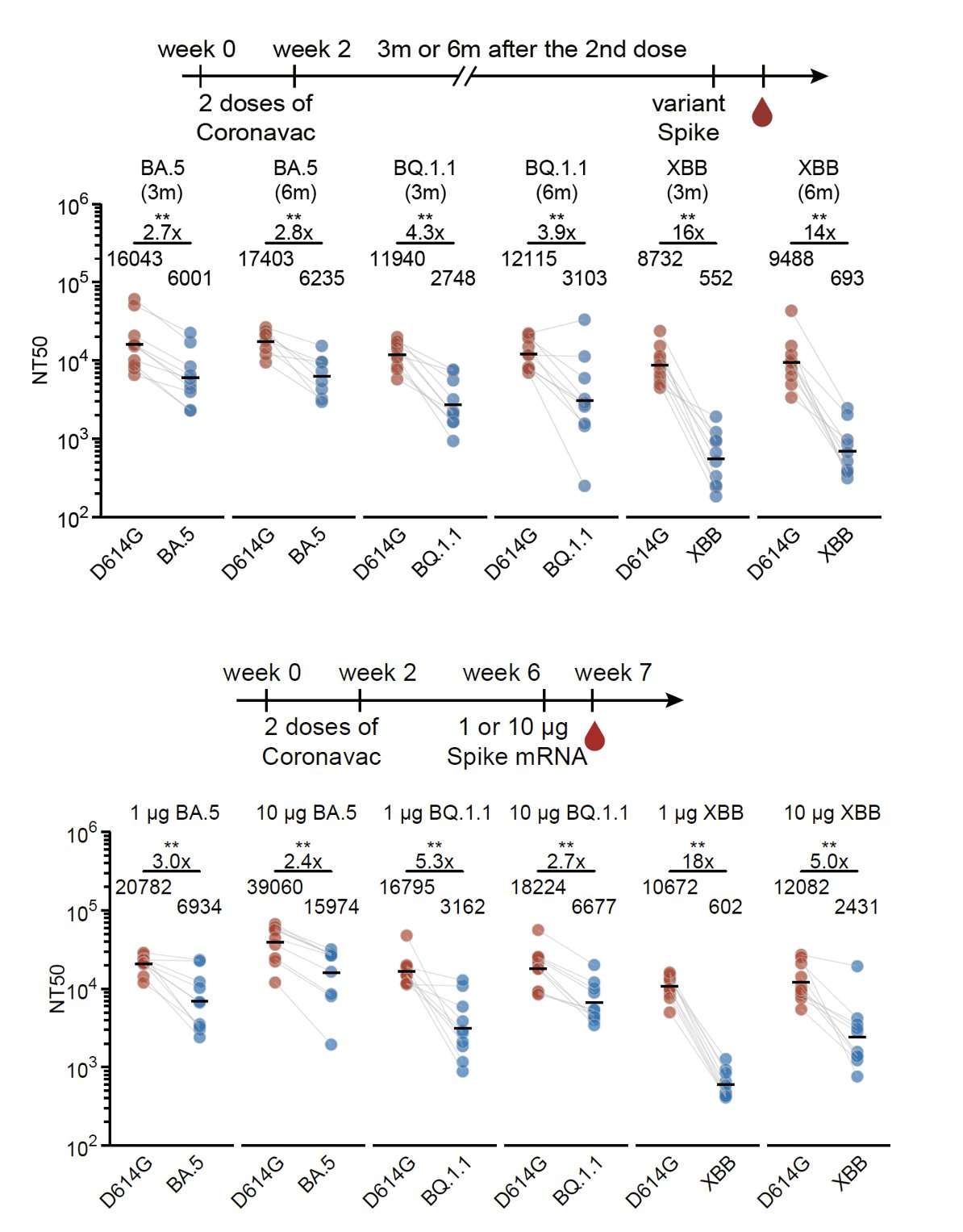
The above data showed the impact of immune imprinting on single Omicron-boosters using variants with long antigenic distances to WT. Nevertheless, we found repeated Omicron boosting significantly ameliorates the situation, especially with prolonged intervals between boosters.
However, we found that two Omicron-boosters after WT priming (4 doses) still generated lower titers compared to corresponding Omicron priming (2 doses) against the boosting variant, suggesting immune imprinting isn't entirely overcome and still limits antibody response.
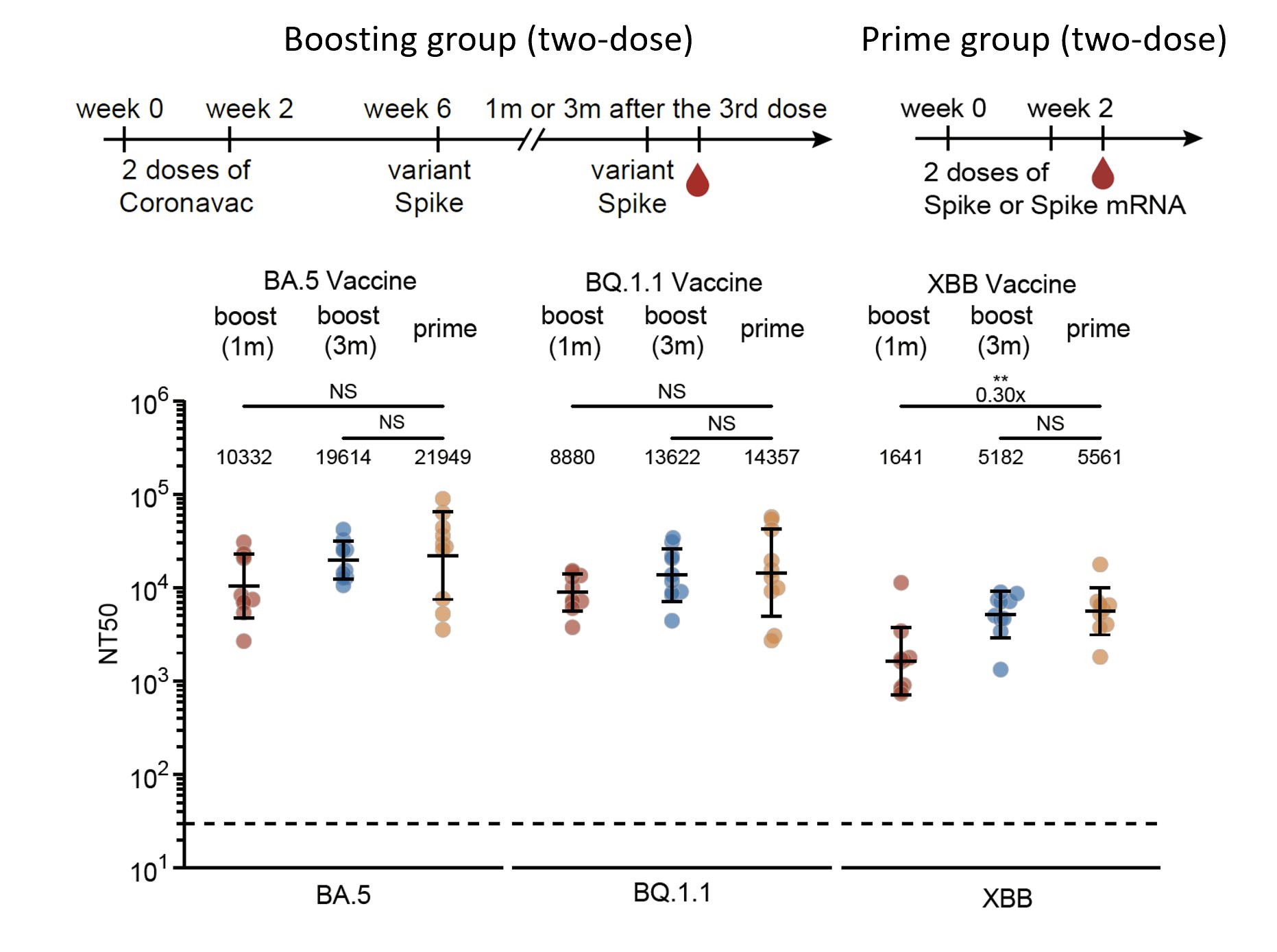
One additional interesting observation is that XBB vaccines demonstrated lower immunogenicity compared to BA.5 and BQ.1.1 in mice. We have confirmed this observation with multiple vaccine manufacturers.
Is our observation in mice applicable to humans? To verify this, we followed individuals with BA.1 breakthrough infections (BTIs) over a year and found that indeed repeated Omicron infections can generate much higher titers across Omicron variants, including XBB.1.5.
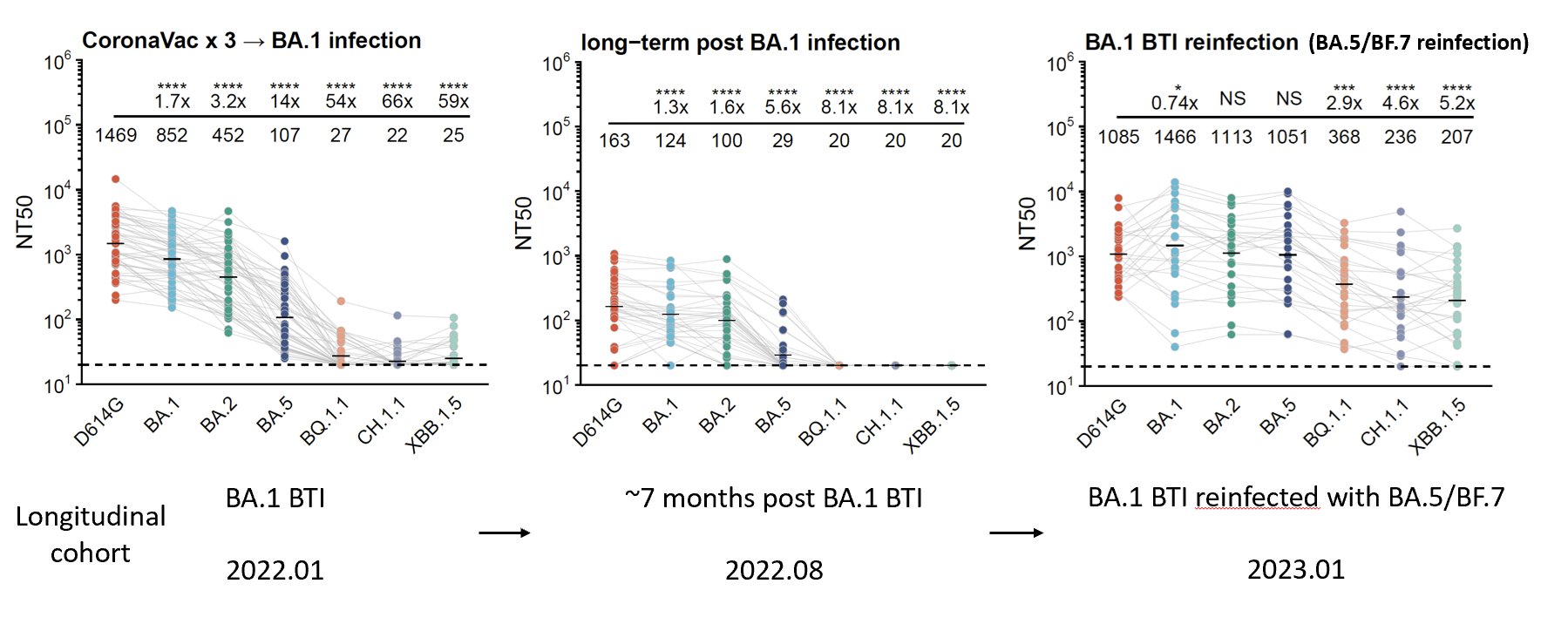
This is also validated in BA.2 BTI cohorts, indicating repeated Omicron infections could alleviate immune imprinting. However, similar to mice, people who got repeated Omicron infections without WT vaccination still generated the highest neutralizing titers across Omicron.
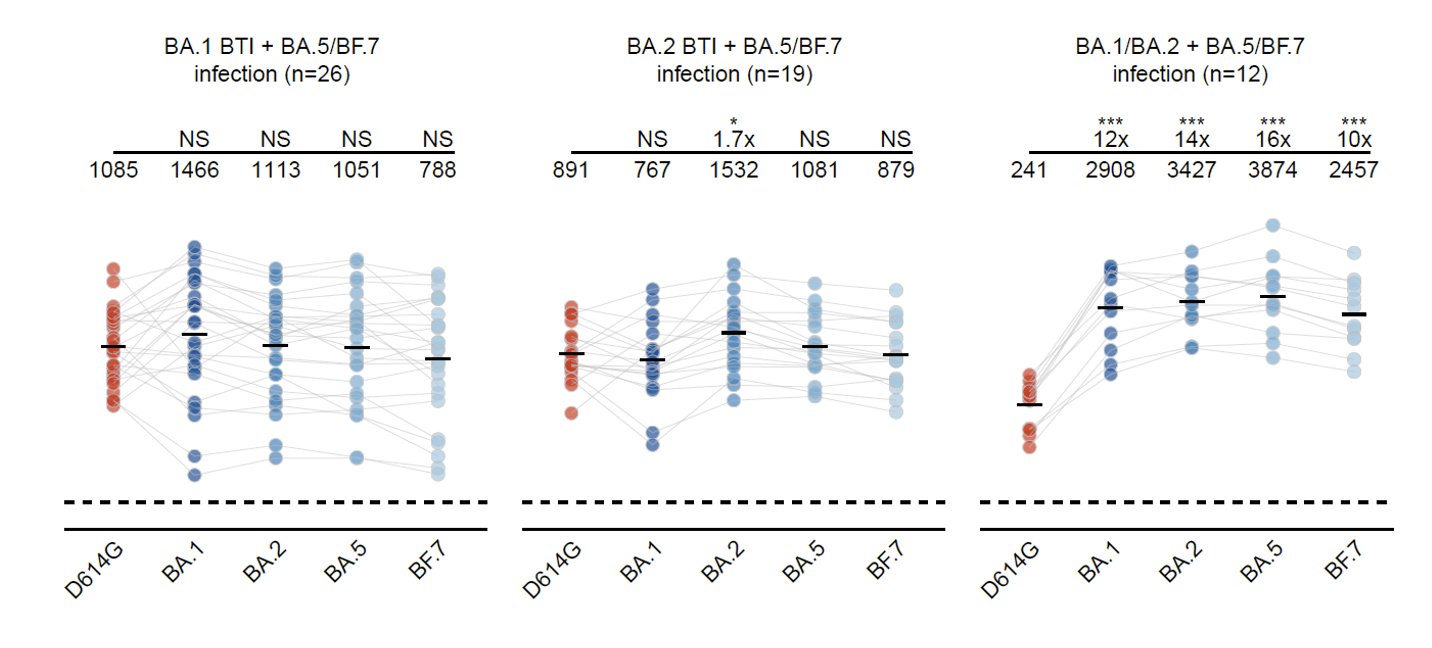
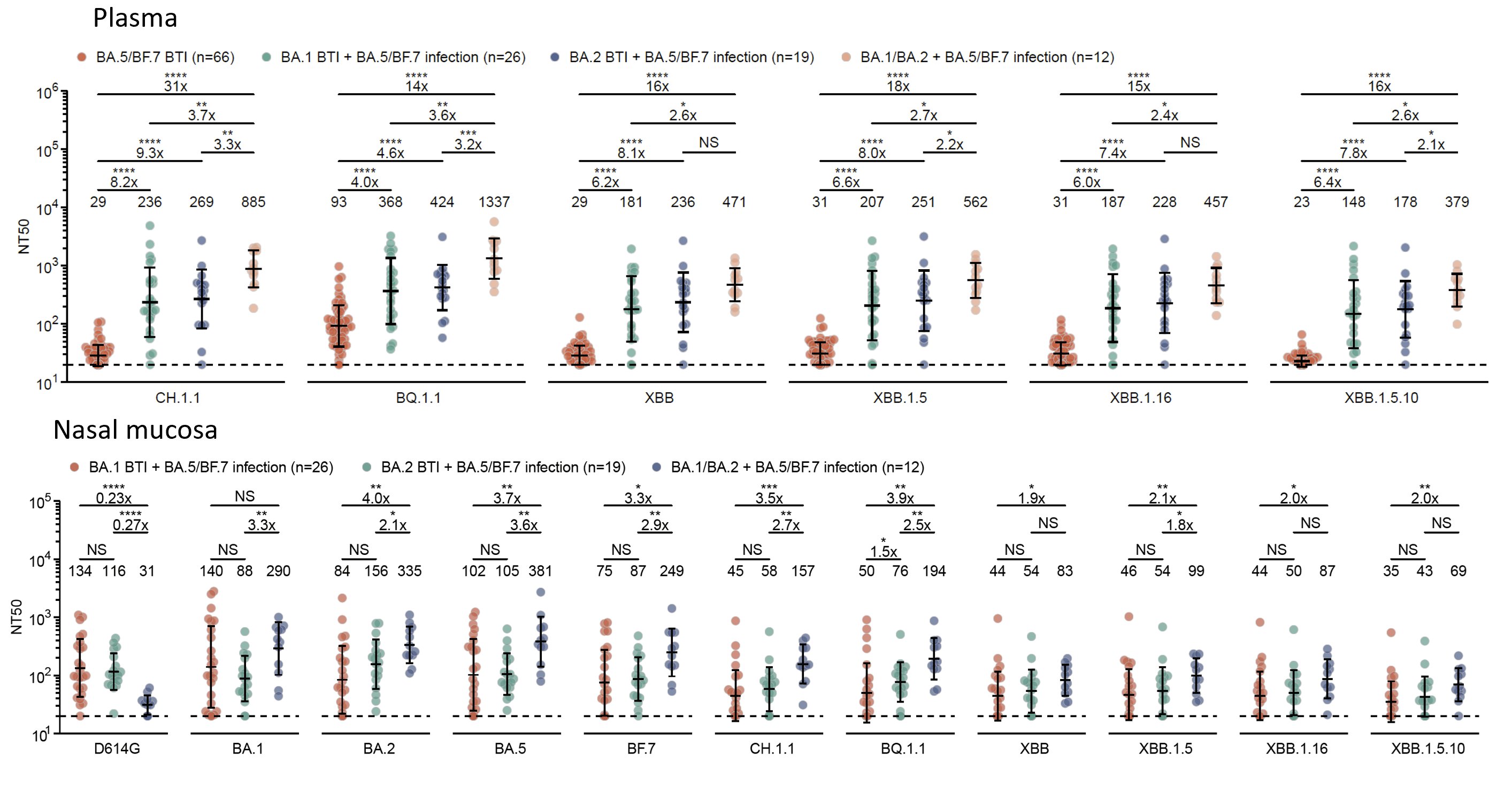
Compared to one-time Omicron breakthrough infections, repeated Omicron infections generated significantly higher neutralization responses toward circulating SARS-CoV-2 variants, including XBB.1.5, XBB.1.16, and XBB.1.5+F456L, in both plasma and nasal mucosa.
Using FACS, we saw an enriched proportion of RBD-binding Omicron-specific memory B cells in cohorts that experienced long-period maturation after BA.1/BA.2 BTI or repeated Omicron infections, which is very different compared to single Omicron BTIs.
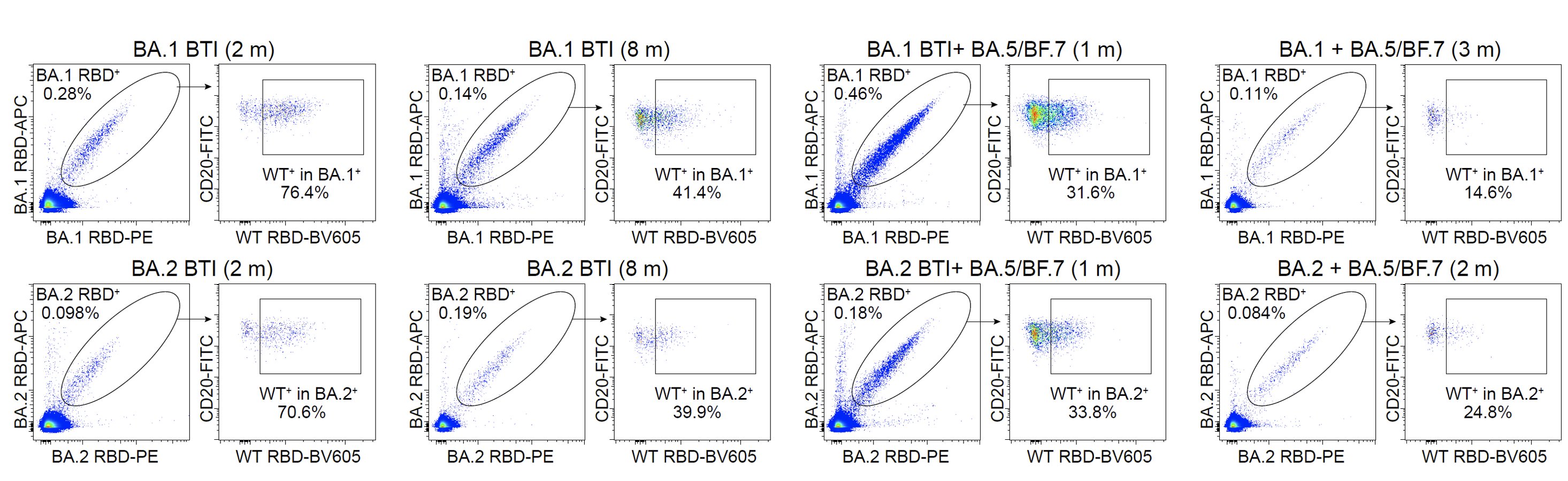
We also isolated those RBD-binding B cells and expressed their encoding antibodies. Indeed, an increase in the proportion and maturation of Omicron-specific antibodies can be observed in repeated Omicron infection cohorts, which explains the alleviation of immune imprinting.
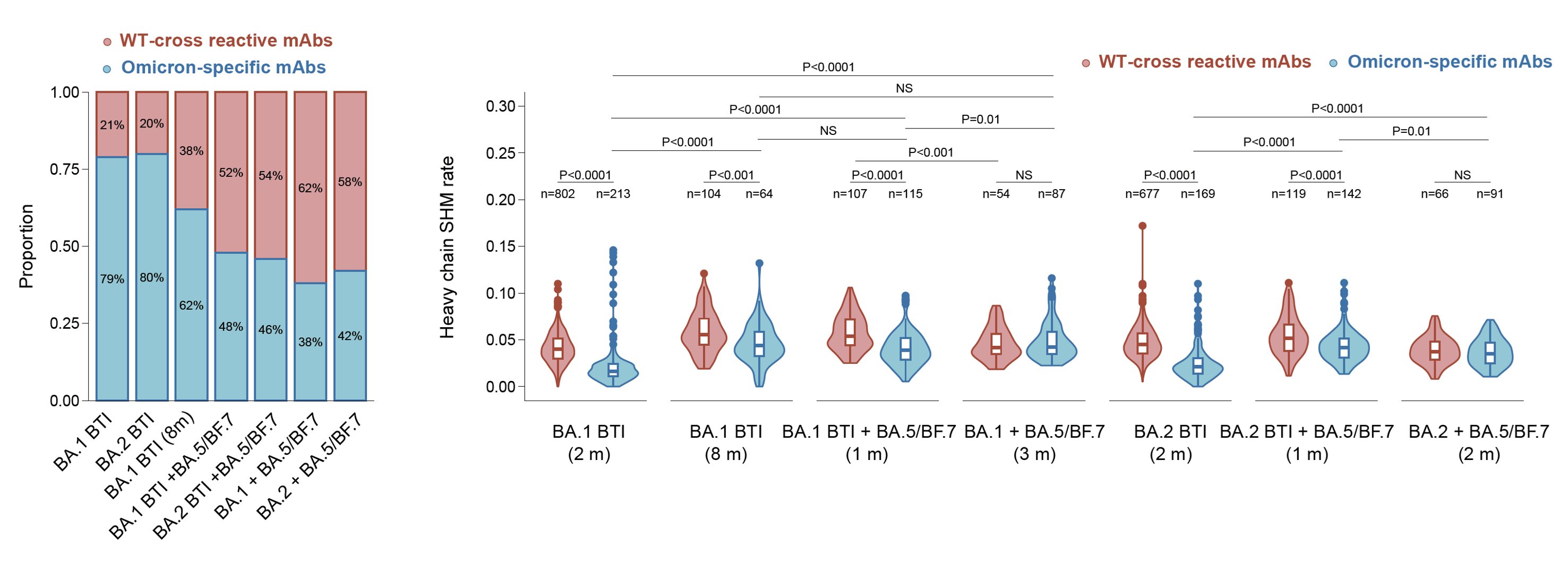
To analyze whether new RBD epitopes are introduced by these Omicron-specific mAbs, we determined their escaping mutation profiles through deep mutation scanning (DMS) based on BA.5-backbone (can't bind WT RBD) alongside previously isolated BA.5-binding mAbs (1350 in total).
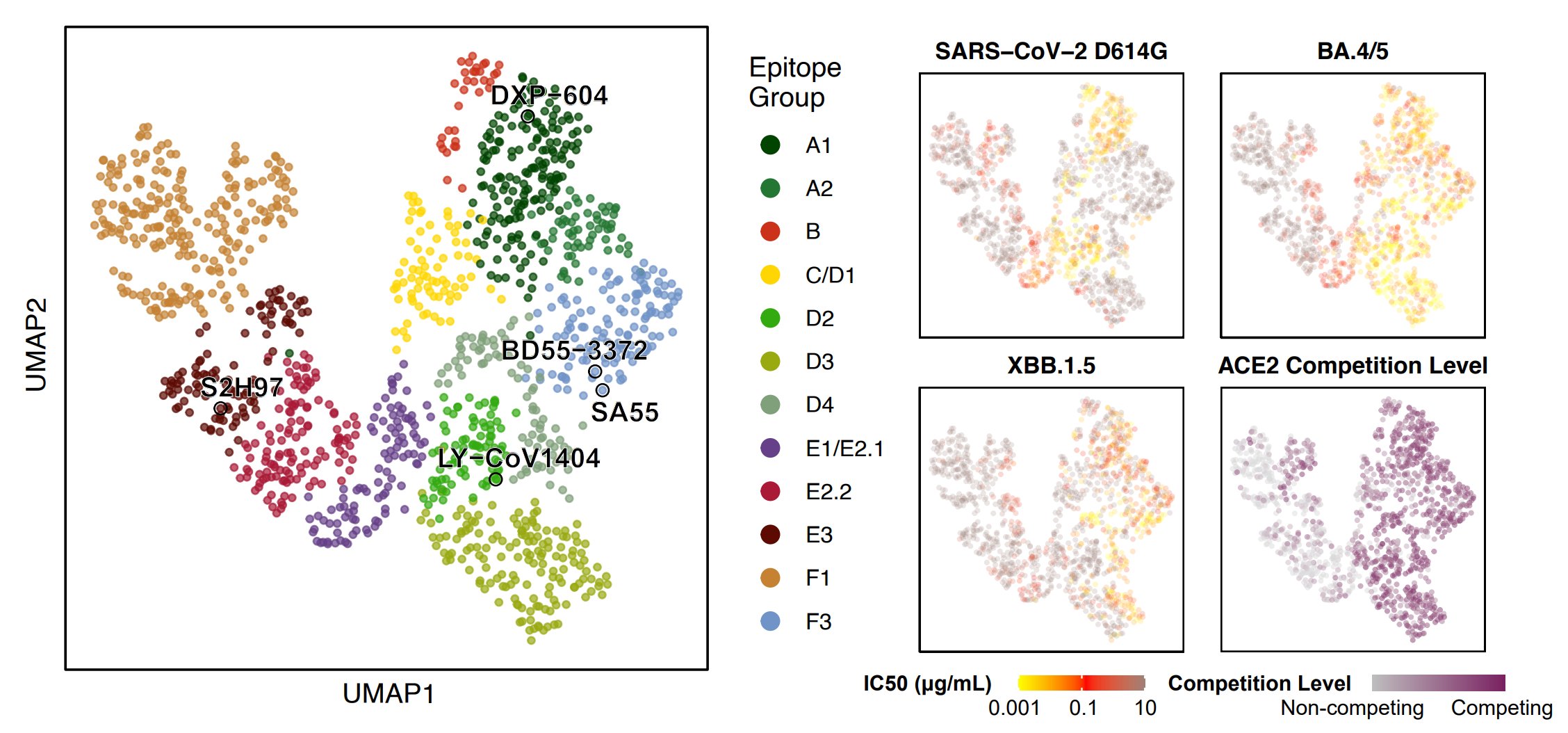
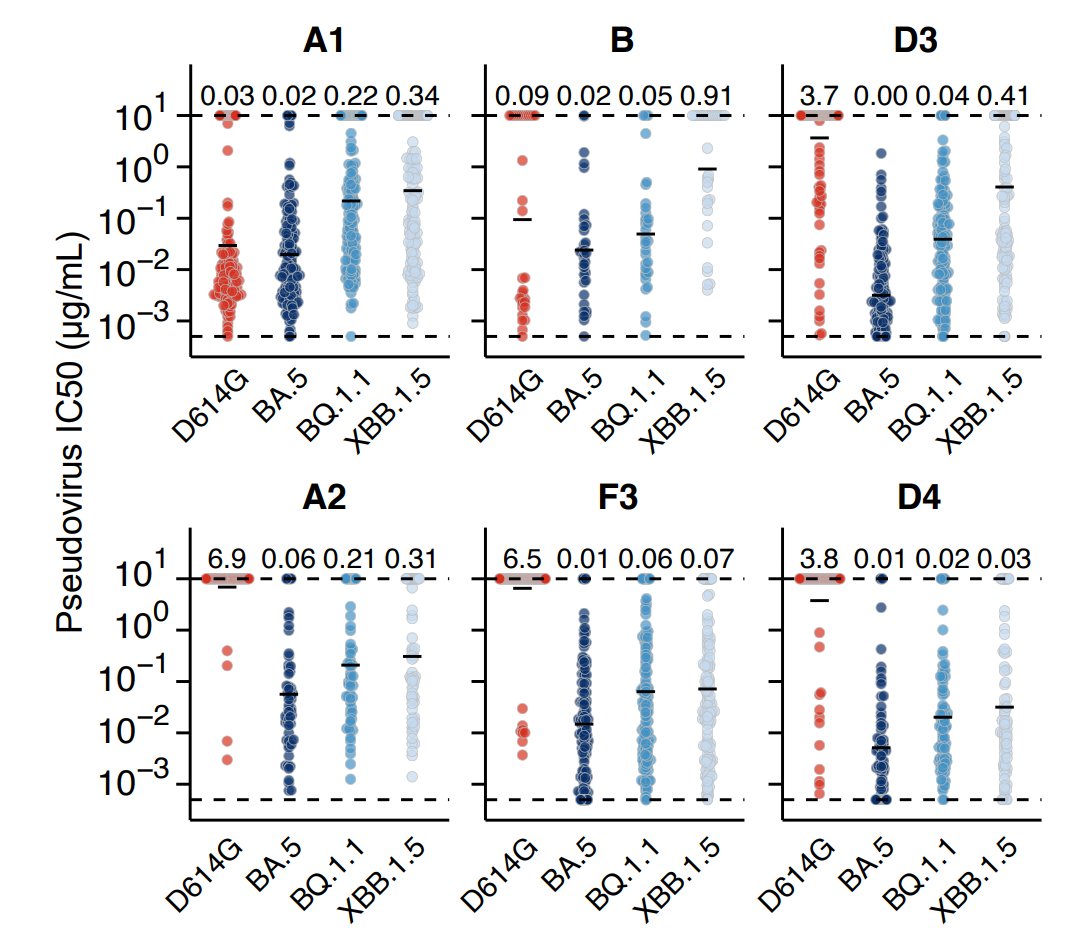
By unsupervised clustering based on the DMS data, we identified some novel RBD epitopes that mainly comprised Omicron-specific mAbs, which we defined as Group A2, D3, D4, and F3. Their epitopes are centered around Omicron mutations N501Y, Y505H, N440K & Q498R.
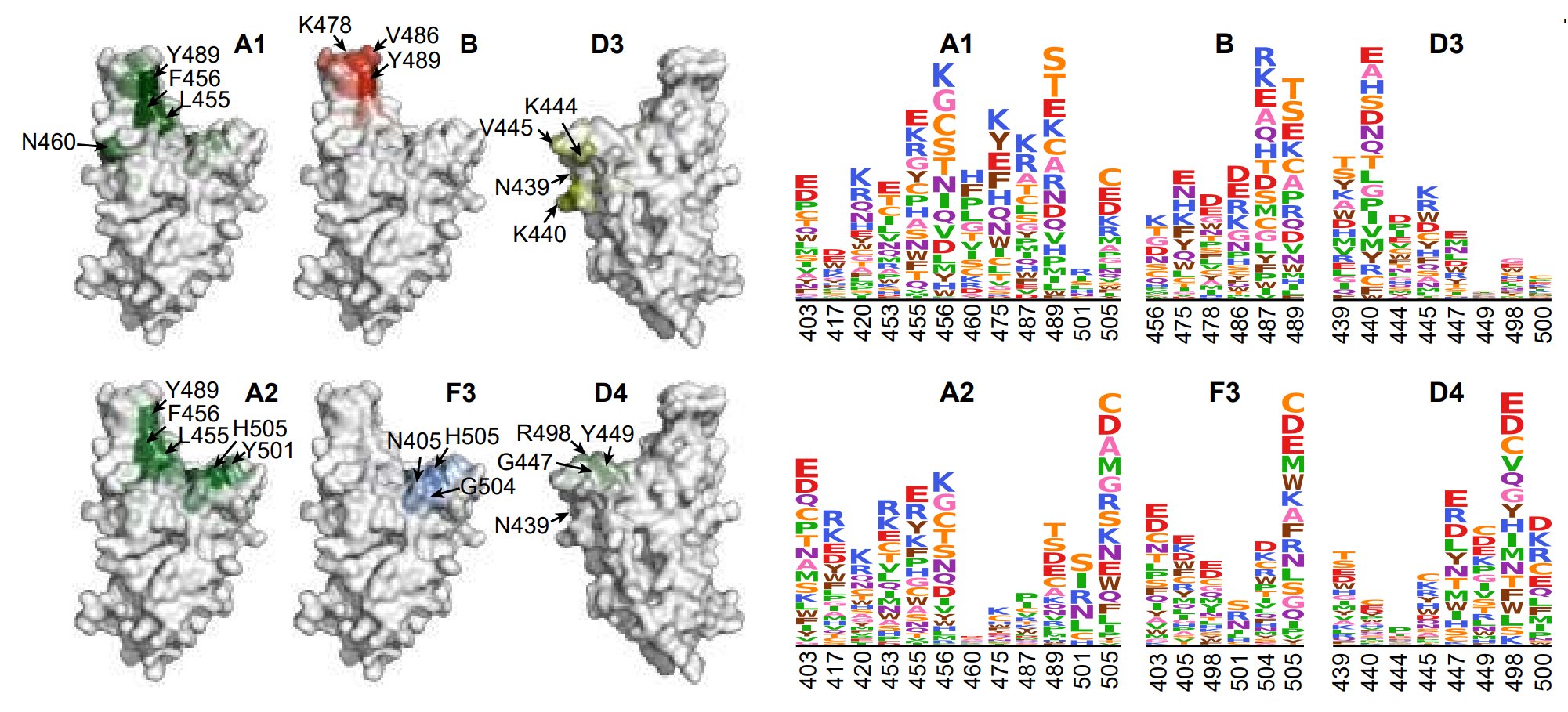
Omicron-specific mAbs in A2, D3, D4 and F3 are enriched with highly potent neutralizing mAbs against BQ.1.1 and XBB.1.5. Notably, most mAbs in F3 here do not cross-react with WT, different from the rare sarbecovirus-neutralizing mAbs in F3 described previously, such as SA55.
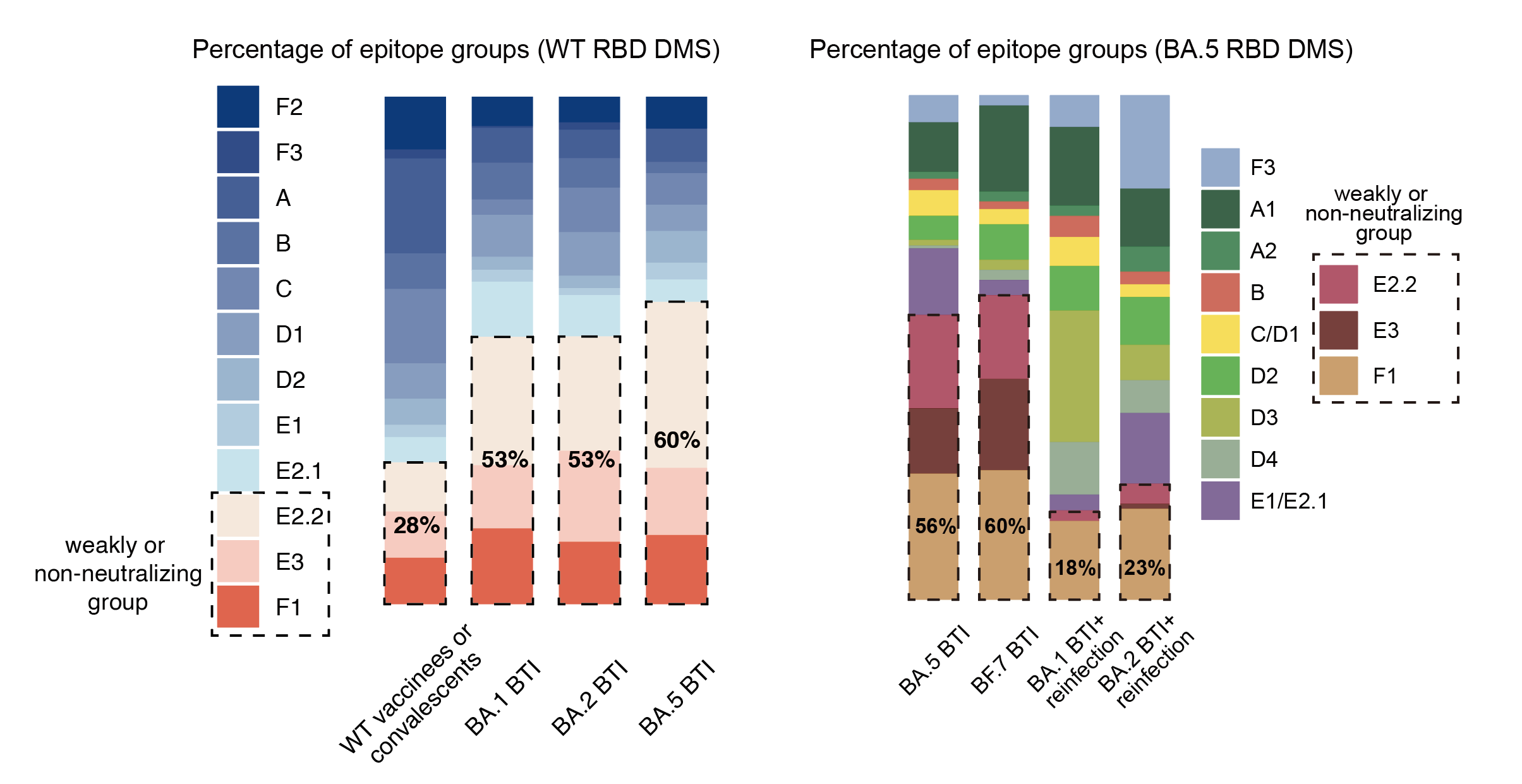
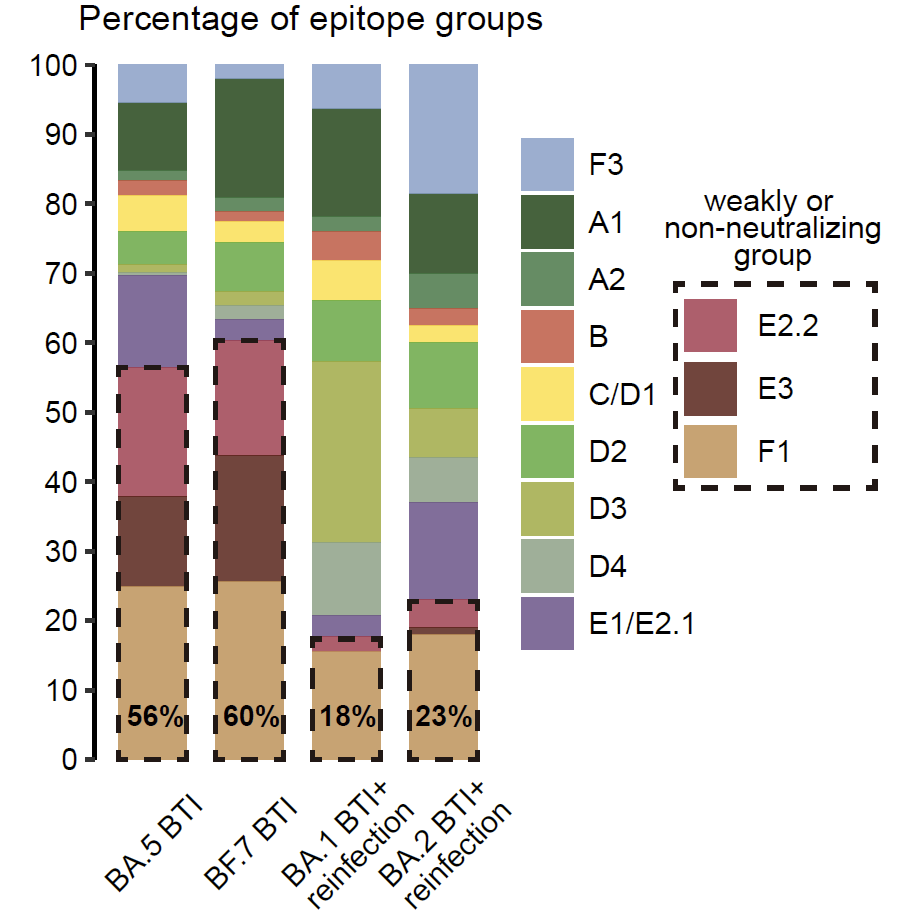
Importantly, consistent with B cell FACS data, single Omicron BTI induces >50% mAbs targeting non or weakly-neutralizing epitopes, while repeated Omicron infection induces only ~20% mAbs targeting such epitopes, indicating striking alleviation of immune imprinting.
Similar to our previous evolution predicting approach, based on the new DMS data, we integratively evaluated the preference of each mutation, considering their impacts on NAb escape, ACE2 binding, and codon constraints and calculated a preference score for RBD mutations.
Here's what the result looks like when inferring the evolution trend for XBB.1.5 RBD only considering mAbs from Omicron BA.5/BF.7 single BTI only. Significant hotspots are R403, N405, N417, Y453, L455, F456, and H505. 403K, 456L, and 453F have already appeared on XBB.1.5.
With NAbs from repeated Omicron infections included, scores of N439, K440, K444, and P445 become higher, corresponding to Group D3 and D4, which are consistent with the enriched epitope distributions of mAbs from each cohort. 445S has started appearing on XBB lineages.
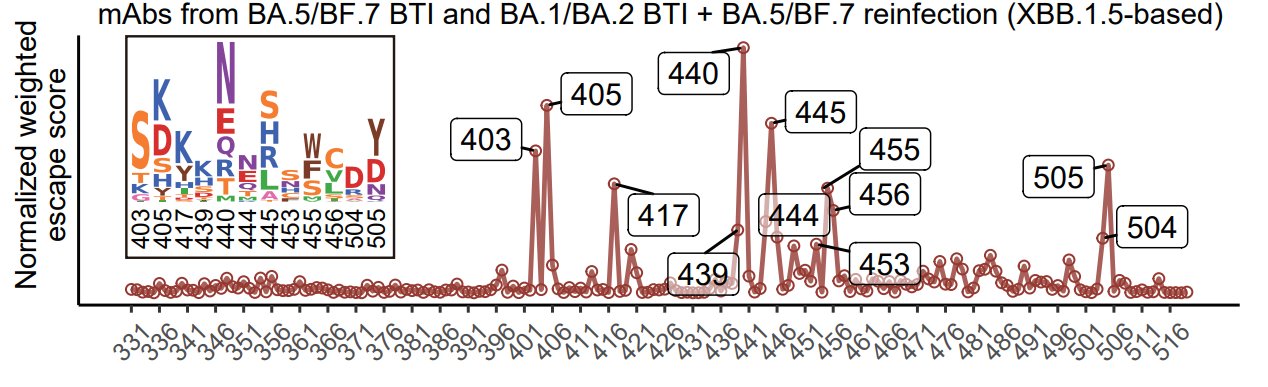
Notably, N405D and N417K reversions should hardly appear due to the potential recovery of previously escaped NAbs. However, K440N and H505Y reversions might appear since the mutations aren't really that immune evasive, so the reversions won't cause noticeable NAb recovery.
K478X is also not identified here due to the low level of 478-sensitive mAbs in our cohort. It's possible that Delta-imprinted convalescents with reinfections generate abundant K478X-sensitive mAbs, as Delta carries T478K. That's why K478X is mostly observed in India.
It's critical to test how these RBD hotspots could combine to achieve max immune evasion without losing affinity to ACE2. After careful consideration, besides the emerging K478R/F456L, we selected 7 additional mutations and sequentially added them to XBB.1.5 pseudovirus.
The 7 mutations are selected from a larger set of mutation candidates considering their impacts on hACE2-binding affinity as determined by surface plasmon resonance (SPR) and @jbloom_lab's DMS data. Y453F and R403K were chosen to maintain high ACE2 binding.
We considered their NAb escaping capability as well, based on the neutralization test of 131 potent XBB.1.5-effective NAbs from 8 epitope groups. F456L, H505Y, K444T, K440N, A484P & N405K are strong mAb-evading mutations that combine together could escape the majority of the panel.
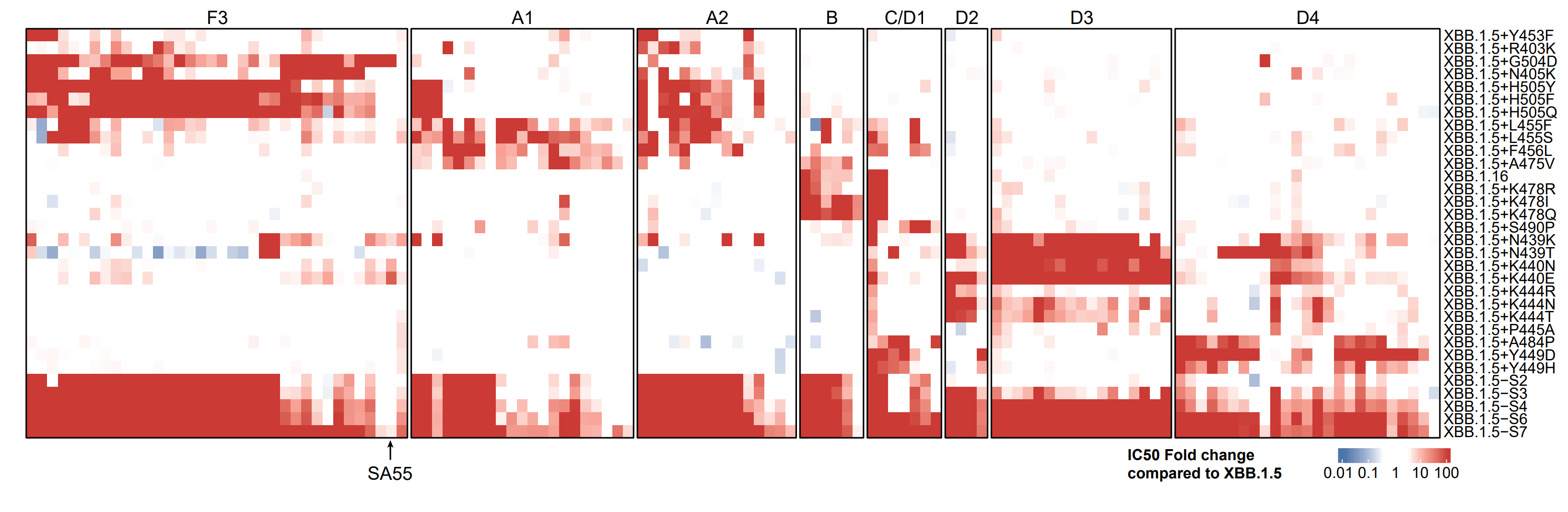
As expected, XBB.1.5-S7 significantly escapes plasma from all tested cohorts, with 505Y&440N contributing mostly. Notably, reinfections with prior BA.1 or BA.2 BTI results in distinct patterns of evasion.
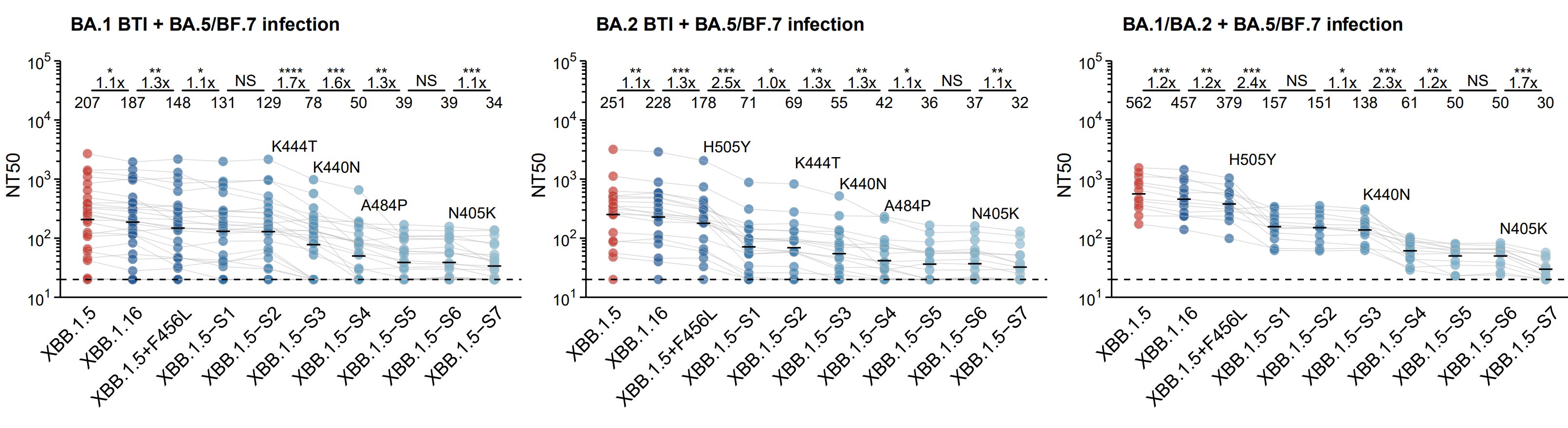
Indeed, mAbs from BA.1 BTI+BA.5 reinfection elicit more D3 mAbs that are weak to 444T/440N, while BA.2 BTI+BA.5 reinfection elicit more F3 and A2 mAbs that are weak to H505Y. This indicates Omicron BTI history also introduces new Omicron-based imprinting.
Notably, K440N and H505Y reversions will not cause substantial recovery of neutralization of WT-reactive NAbs, as verified by testing plasma from BA.5/BF.7 single BTI.
In sum, we demonstrated that matured Omicron-specific mAbs are critical to the alleviation of WT vaccination-induced immune imprinting, and repeated Omicron stimulation would generate a high proportion of them.
These Omicron-specific NAbs target new RBD epitopes compared to WT-induced mAbs and exhibit potent activity across Omicron lineages, including XBB.1.5. They will soon become the major viral immune pressure contributor, and new mutations may emerge as we have predicted.
Our findings also suggest the WT component should be abandoned when updating COVID-19 vaccine antigen compositions to XBB lineages, and those who haven't been exposed to Omicron yet should receive two updated vaccine boosters.
The updated BA.5-based DMS data, analyzed mainly by @jianfcpku, will be shared via @jbloom_lab's online calculator very soon for people to play with. This work is largely contributed by our talented graduate students Ayijiang Yisimayi, Weiliang Song, Jing Wang, and Fanchong Jian (See Members).

