Perspectives on emerging SARS-CoV-2 BA.2.86
This is a copy of our previous thread on Twitter.
Our results on BA.2.86 has now been published on Lancet Infectious Diseases.
Recently, a new saltation SARS-CoV-2 lineage BA.2.86 has emerged and designated as a variant under monitoring (VUM)
by WHO. Here, we would like to share our latest experiment results on this variant. Generally, we found that,
(1) BA.2.86 is antigenically distinct compared to XBB.1.5.
(2) BA.2.86 can significantly escape XBB-infection/vaccination induced antibodies.
(3) However, the infectivity of BA.2.86 may be much lower than XBB.1.5 and EG.5.
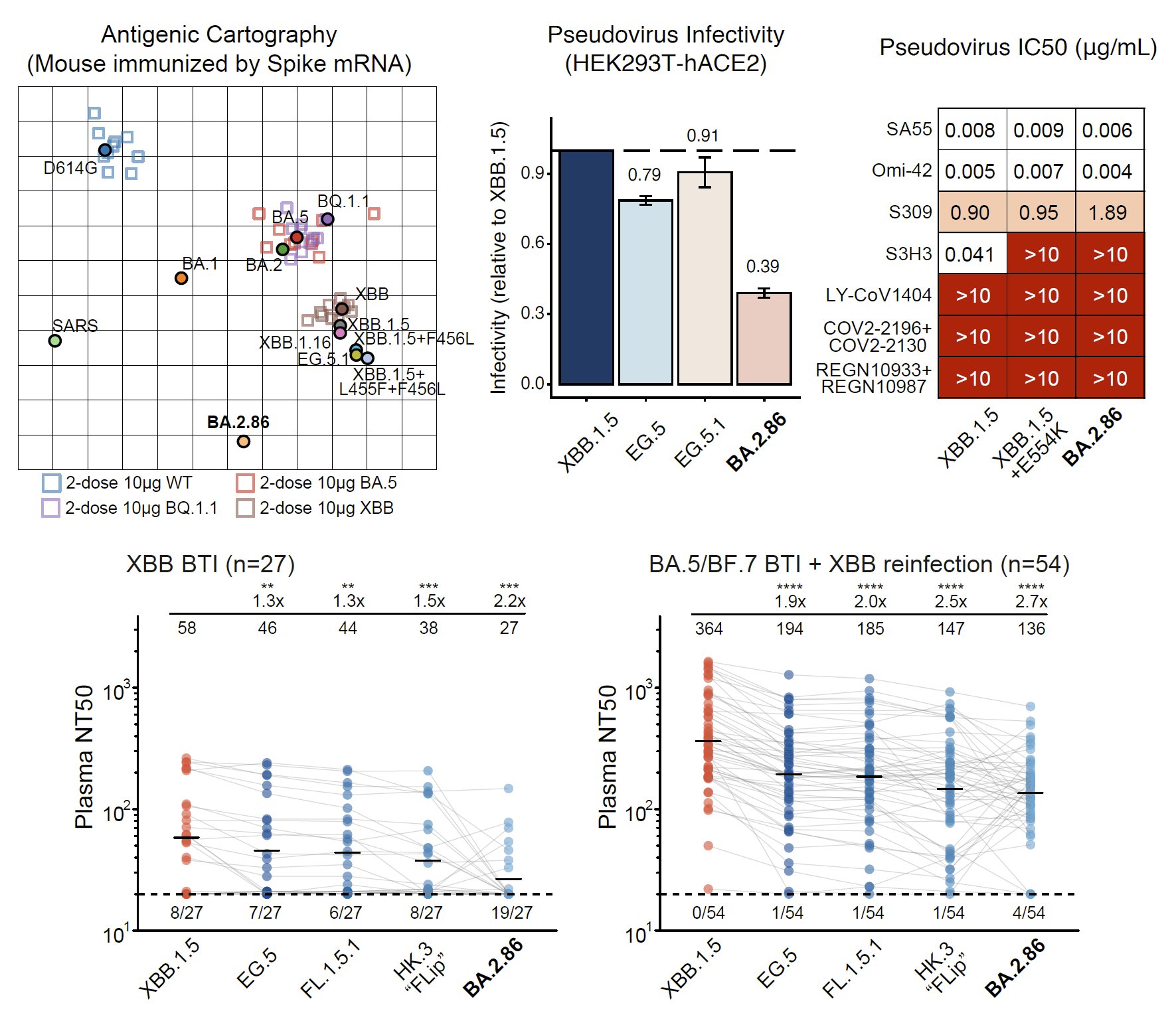
By using pseudovirus neutralization assay and antigenic cartography (based on mRNA immunized mouse serum), we found that BA.2.86 is antigenically distinct from WT, BA.2, BA.5, and XBB.1.5. This means that XBB-induced antibodies cannot well recognize and neutralize BA.2.86.
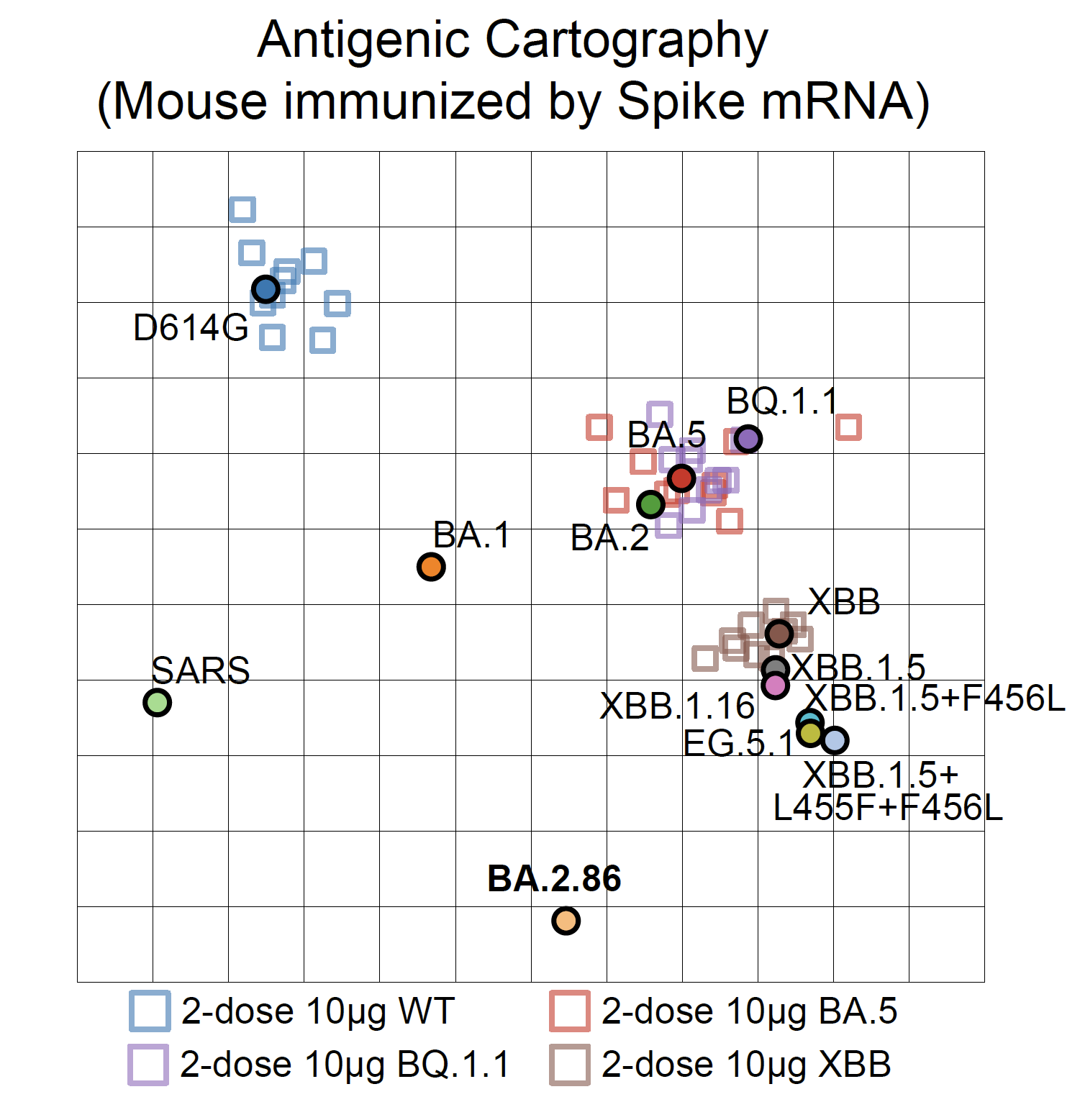
Indeed, BA.2.86 can induce significant antibody evasion of plasma isolated from convalescents who experienced XBB breakthrough infection or reinfections. BA.2.86's immune evasion capability even exceeds EG.5 and is comparable to "FLip" variants (XBB.1.5 + L455F & F456L).
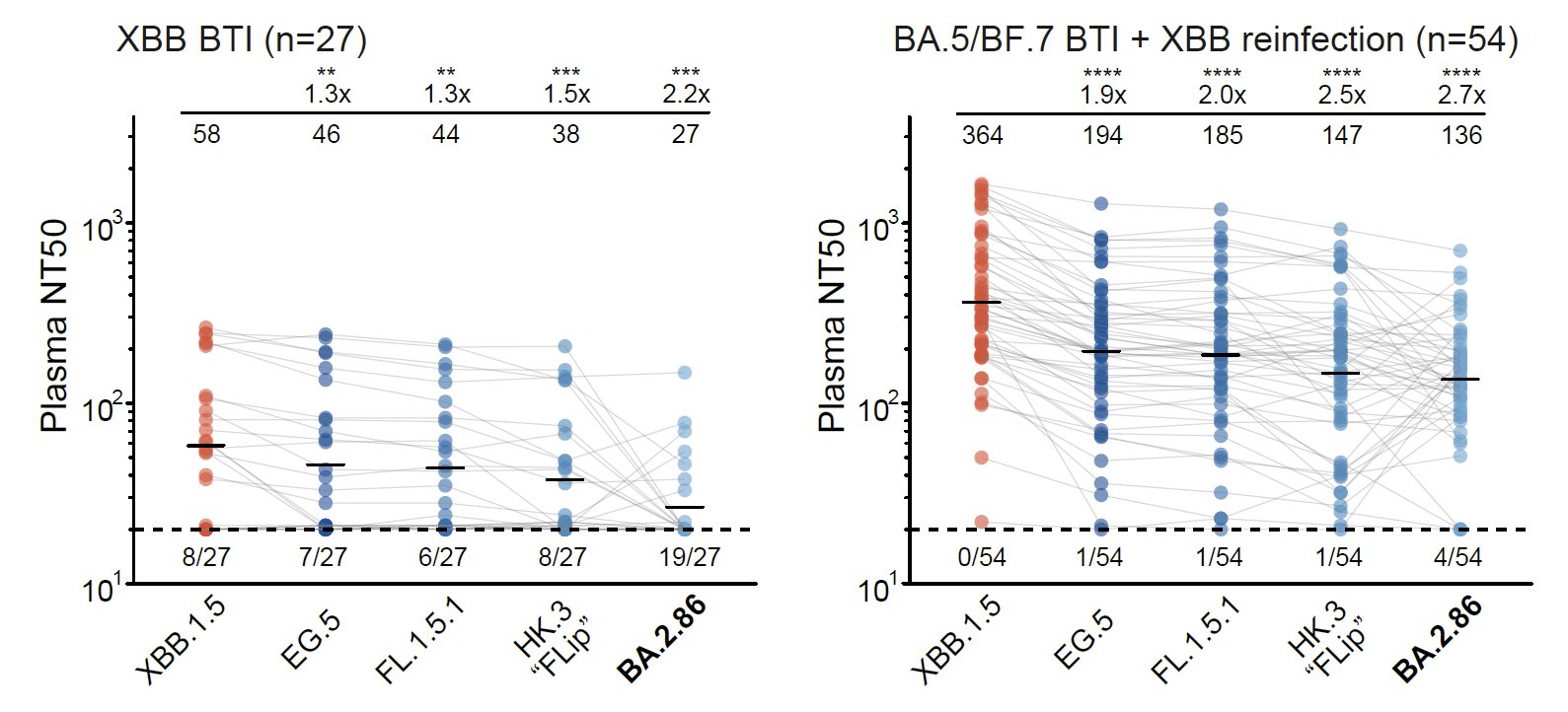
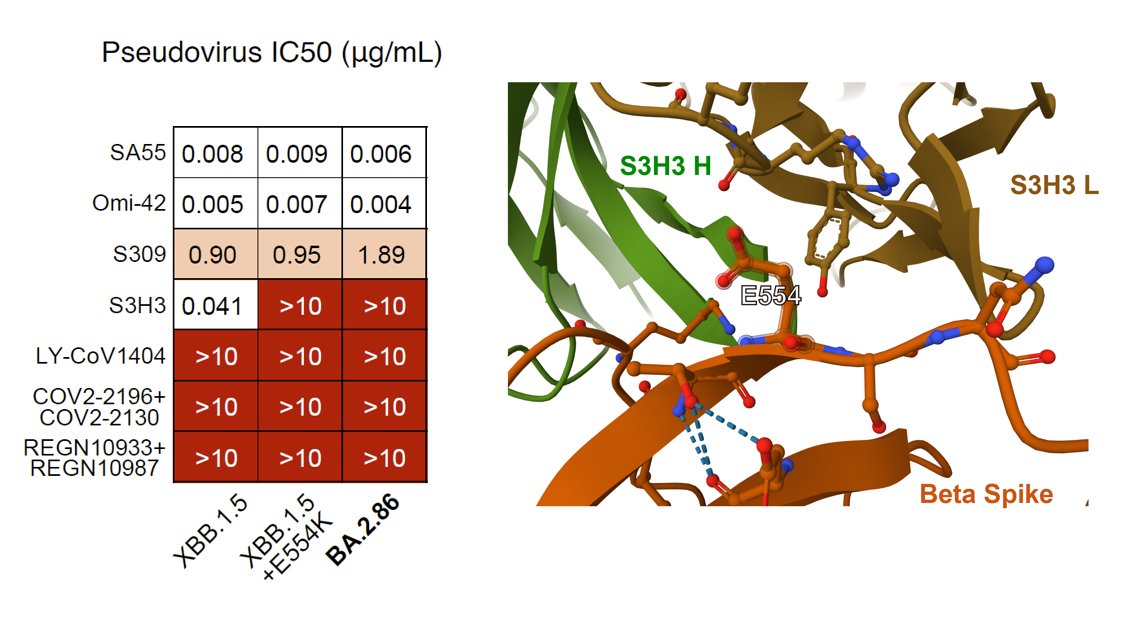
As for monoclonal neutralizing antibody (NAb) drugs, all approved antibodies can't neutralize BA.2.86 well, but SA55 remains effective (clinical phase II-III). Interestingly, the E554K mutation carried by BA.2.86 can escape SD1-targeting NAb, represented by S3H3.
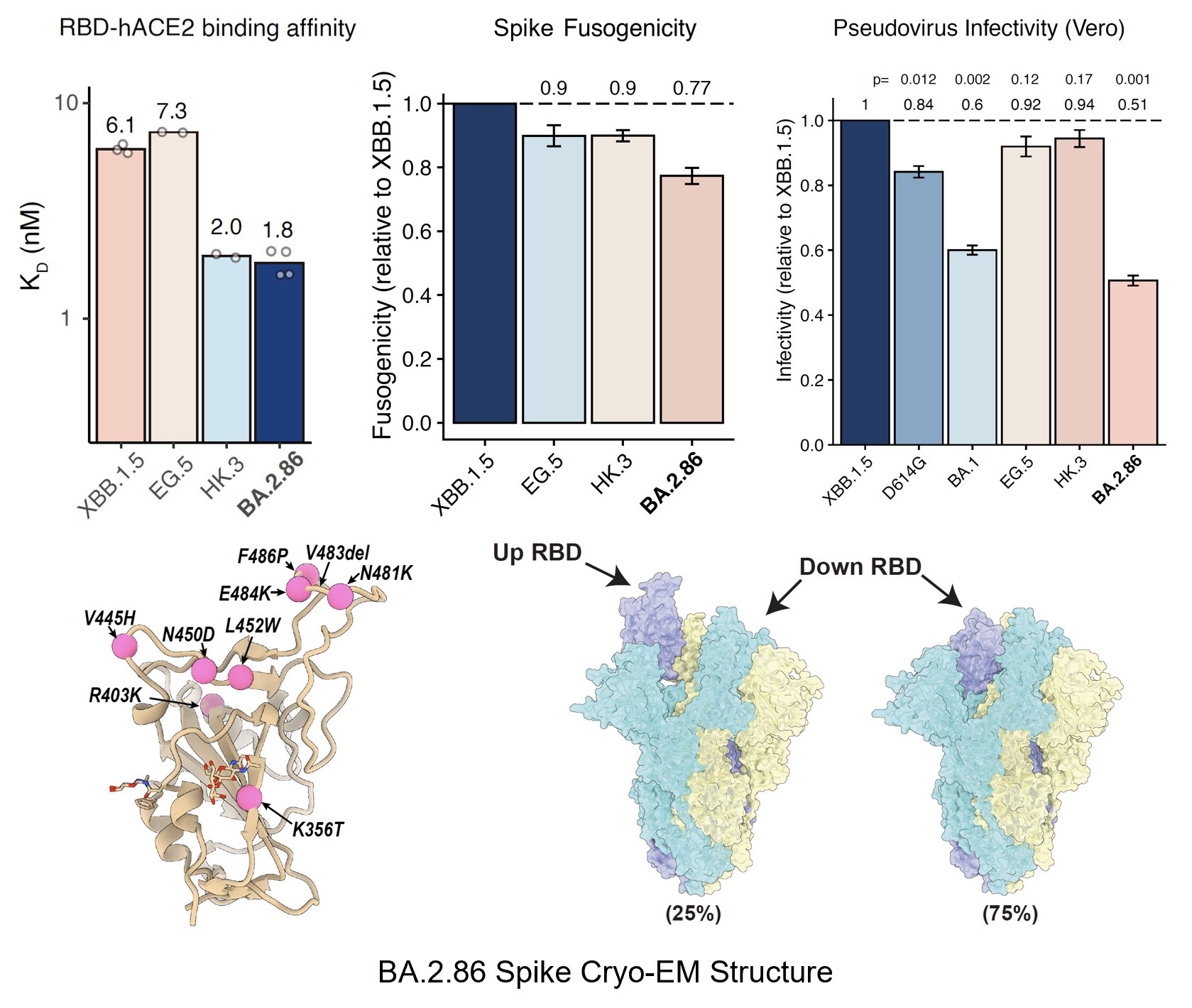
We also tested a panel of XBB.1.5-effective NAbs against XBB.1.5-based pseudoviruses carrying single BA.2.86 RBD mutations. Results showed that N450D, K356T, L452W, A484K, V483del, V445H are the key mutations responsible for BA.2.86's enhanced immune evasion than XBB.1.5.

Importantly, we found that BA.2.86 exhibits lower cell infectivity compared to XBB.1.5 and EG.5, which may affect its transmission. The lower infectivity may be contributed by K356T, V483del, and E554K. Note that here the infectivity is measured through pseudovirus assays.
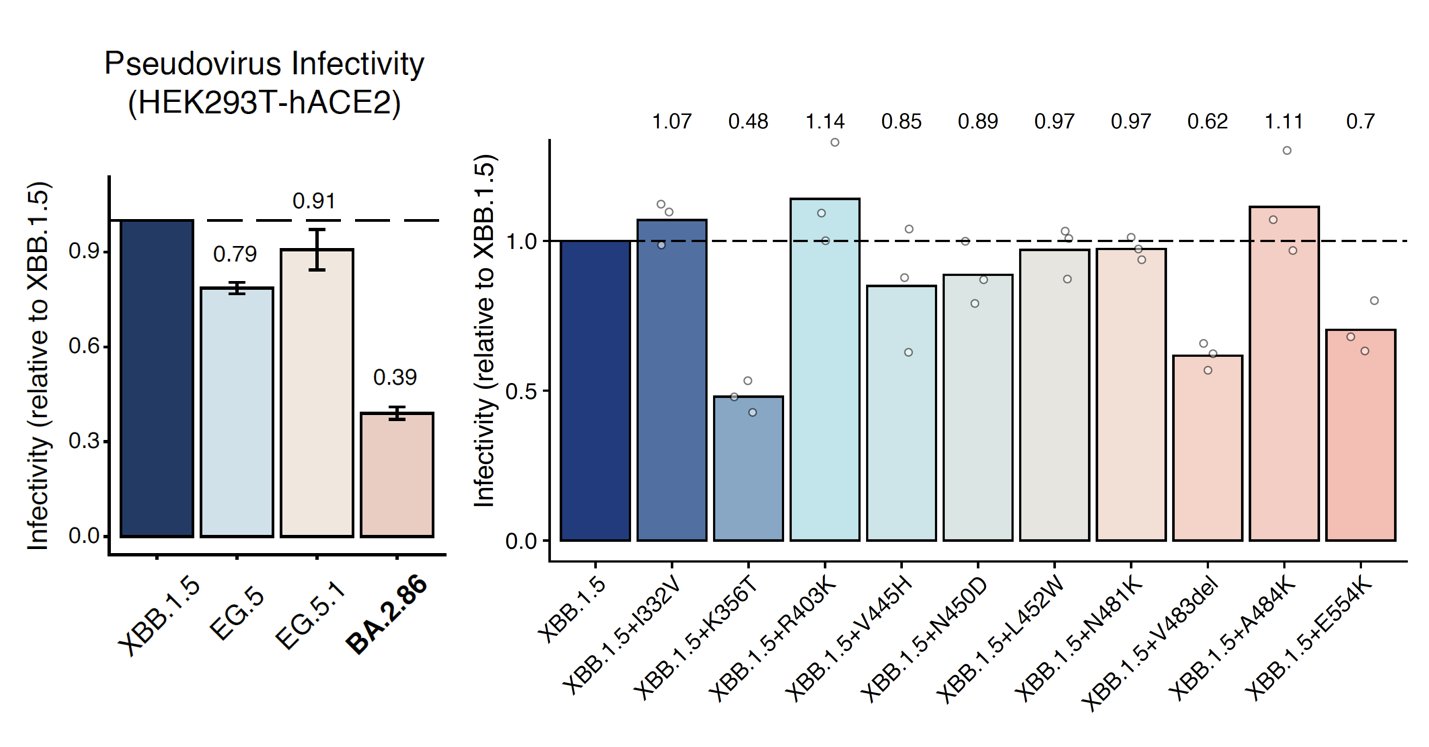
In sum, BA.2.86 is antigenically distinct from XBB.1.5 and can escape XBB-induced neutralizing antibodies. The updated vaccine's efficacy against BA.2.86 should be closely monitored; however, BA.2.86 may not prevail very fast due to its lower infectivity.
The following contents are from anothor thread posted on Twitter.
We further tested the hACE2-binding affinity and structural features of BA.2.86 and found
(1) BA.2.86's ACE2 binding affinity is very high.
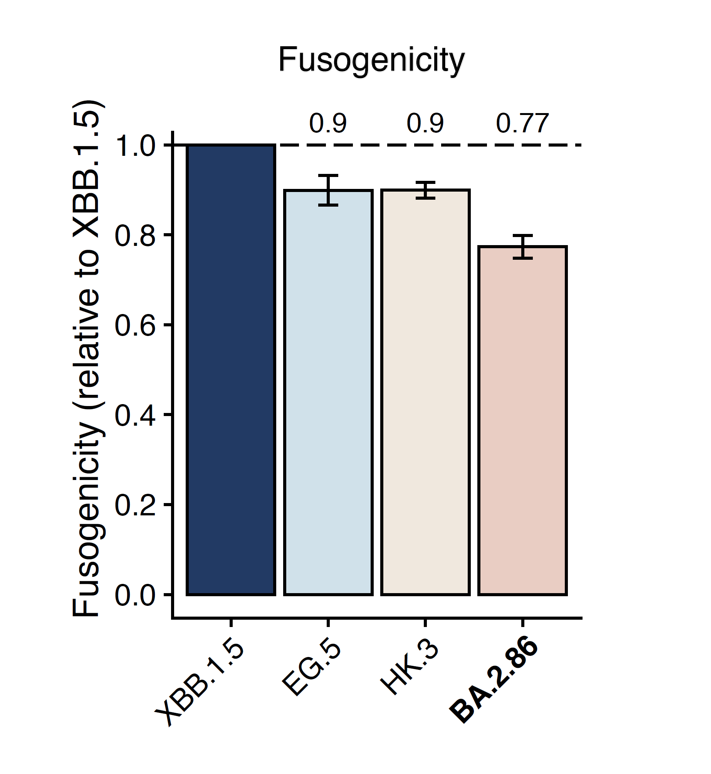
(2) BA.2.86 has lower fusogenicity than XBB.1.5.
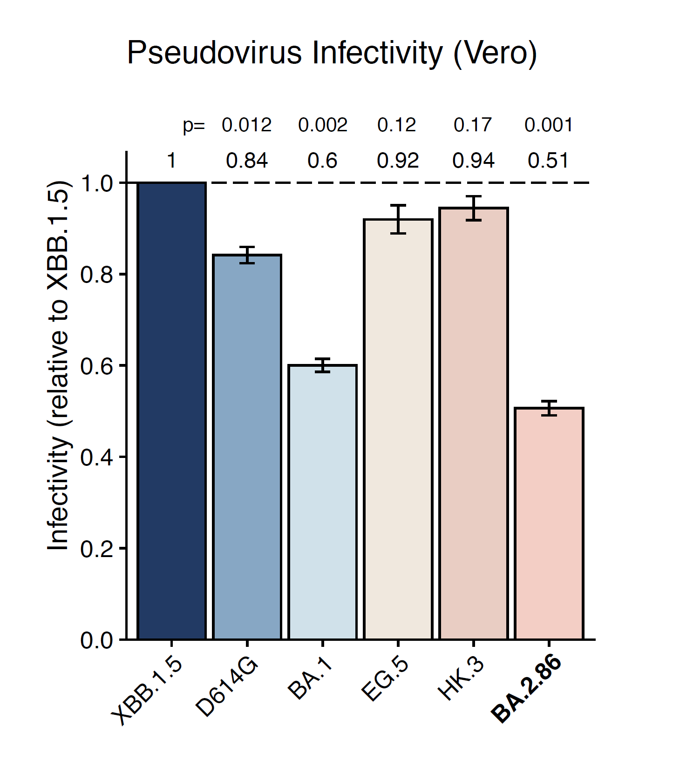
(3) BA.2.86's infectivity in Vero cells is similar to BA.1, lower than XBB.1.5.
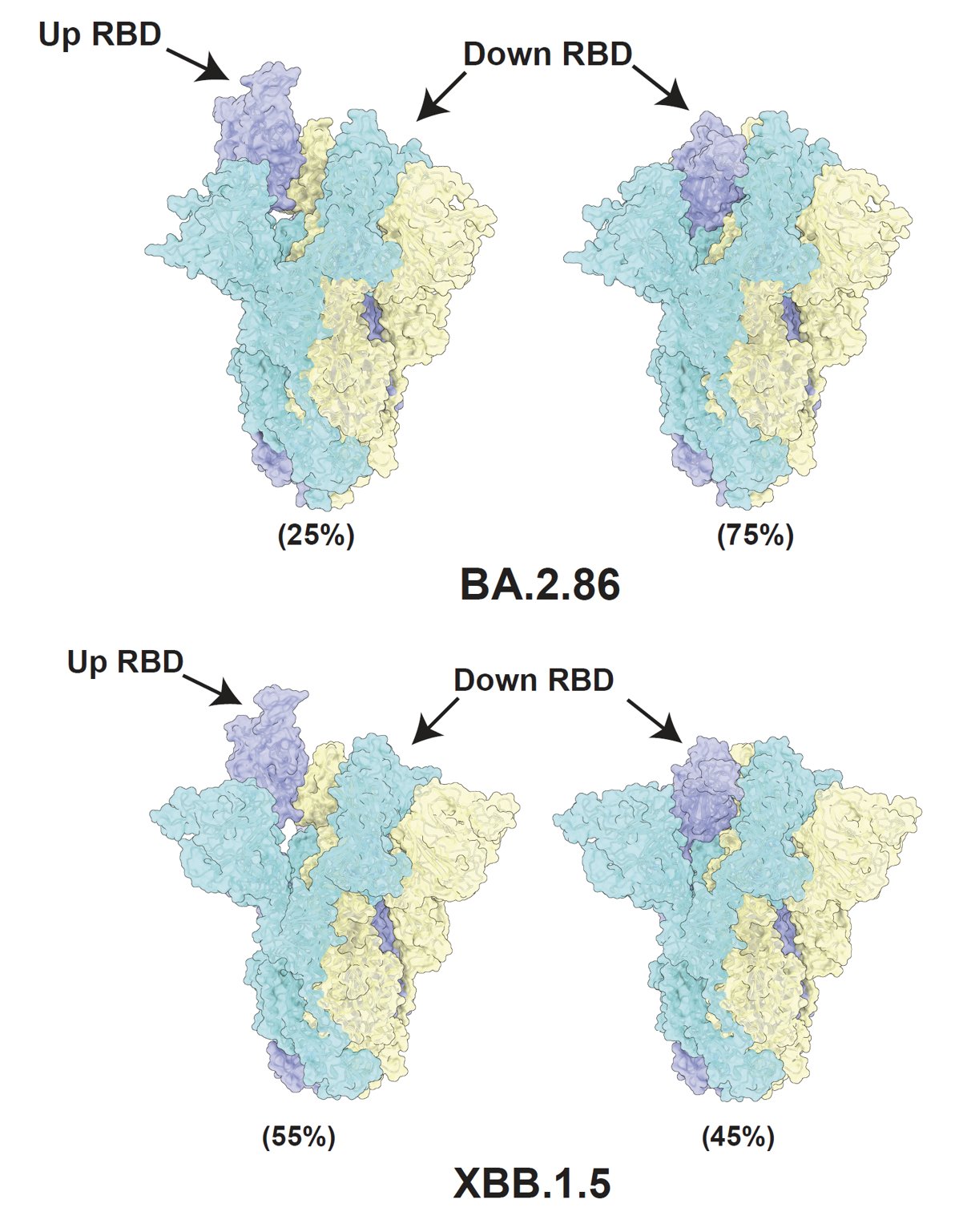
(4) Structure analysis shows that BA.2.86's Spike prefers RBD "down" conformation.
BA.2.86's RBD showed a pretty high hACE2 binding affinity measured by SPR, higher than that of XBB.1.5 and EG.5 and is even comparable to "FLip" variants like HK.3. BA.2.86's V483del indeed decreases ACE2 binding, but R403K is just too powerful and makes up for the loss.
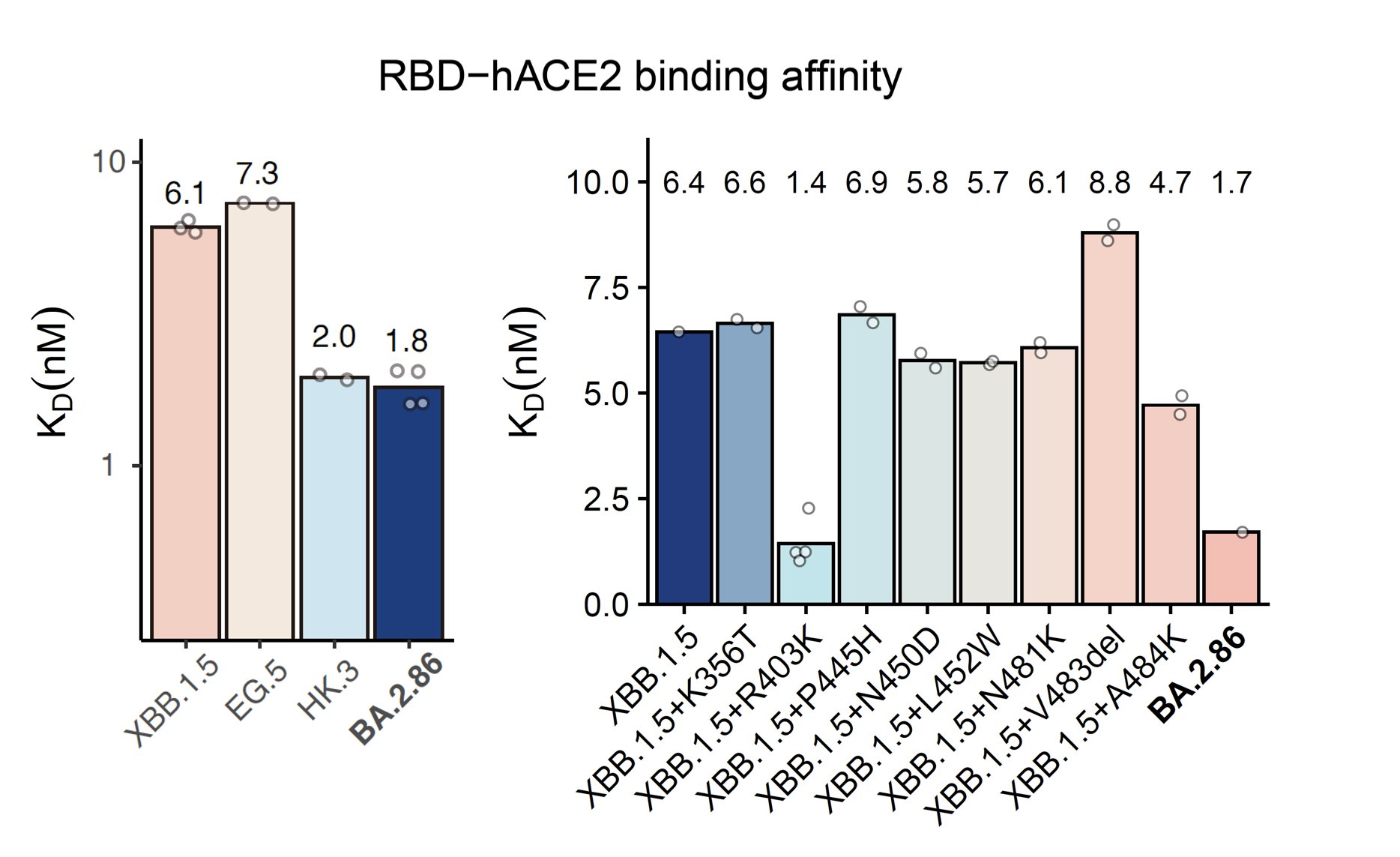
We also measured the cell-cell fusion capability using Spike-transfected 293T cells and 293T-hACE2 cells. BA.2.86 showed a lower fusogenicity than XBB.1.5, despite the fact that BA.2.86's ACE2 binding affinity is much higher. Note this assay is free of pseudoviruses.
We also performed the pseudovirus infection assay in Vero cells and found BA.2.86's infectivity is still lower than XBB.1.5, and is comparable to BA.1. Since BA.1 can transmit pretty well, this suggests BA.2.86 can also prevail as long as its immune evasion is strong enough.
To investigate why BA.2.86 has higher ACE2 binding but lower infectivity and fusogenicity, Xiangxi Wang's lab helped us solved the cryo-EM structure of BA.2.86's spike. Surprisingly, most of BA.2.86's spike has its RBD in "down" position compared to XBB.1.5.
Favoring "down" RBDs may explain BA.2.86's lower fusogenicity and infectivity observed in vitro since only "up" RBDs can bind to ACE2. It also suggests that BA.2.86's mutation on SD1-SD2 (where D614G is located) may have affected its spike conformation or spike flexibility.
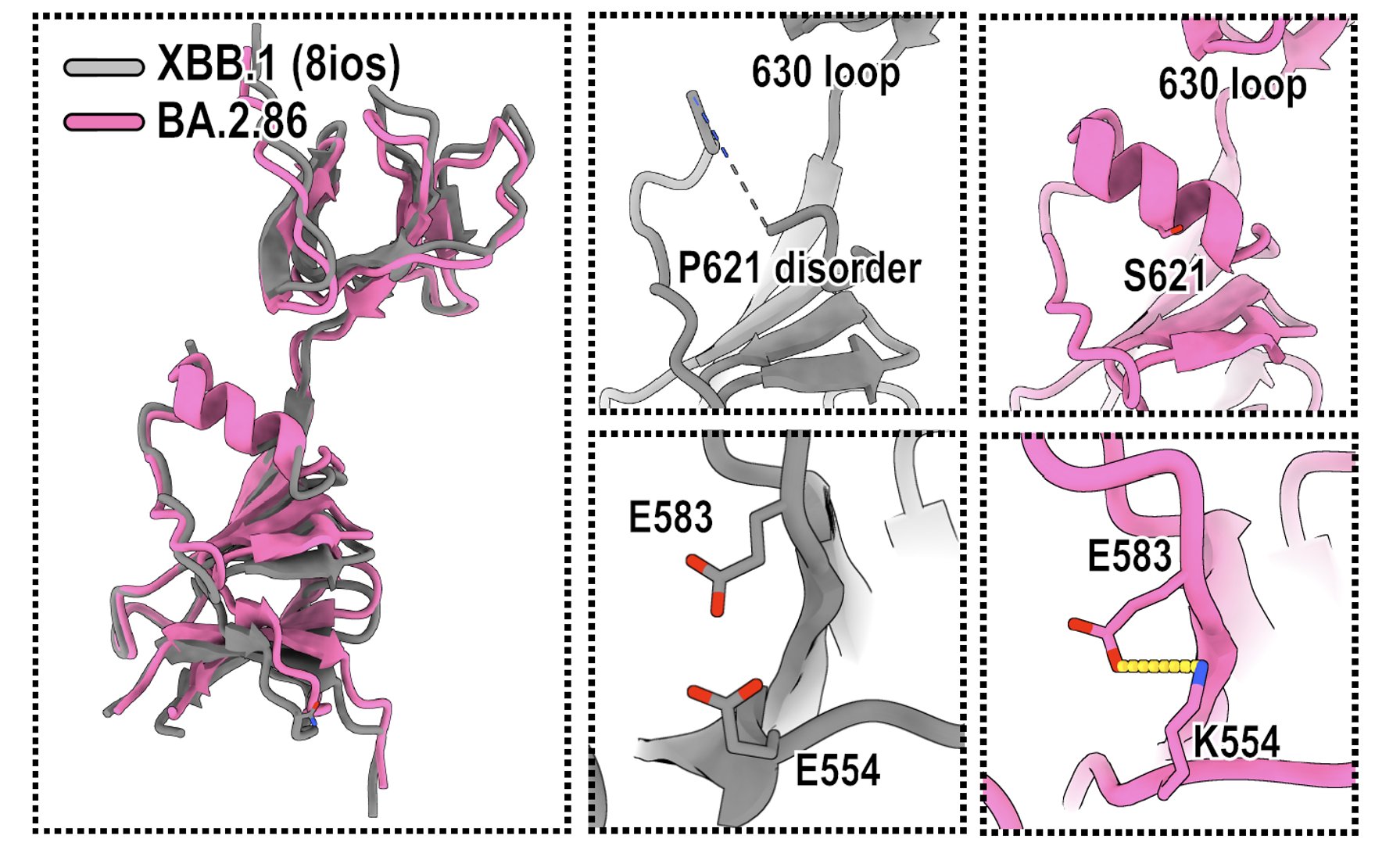
Indeed, BA.2.86's P621S mutation actually created a alpha-helix near the 630 loop, which could enhance the local stability. Also, BA.2.86's E554K mutation formed a salt bridge with 583E, which could also increase stability. These changes may greatly affect spike flexibility.
Some other interesting features of BA.2.86's spike include the additional N354 Glycan introduced by K356T mutation (expected), and that the V483 deletion does not disrupt the C488-C480 disulfide bond, which is the reason why BA.2.86's RBD can still bind well to ACE2.
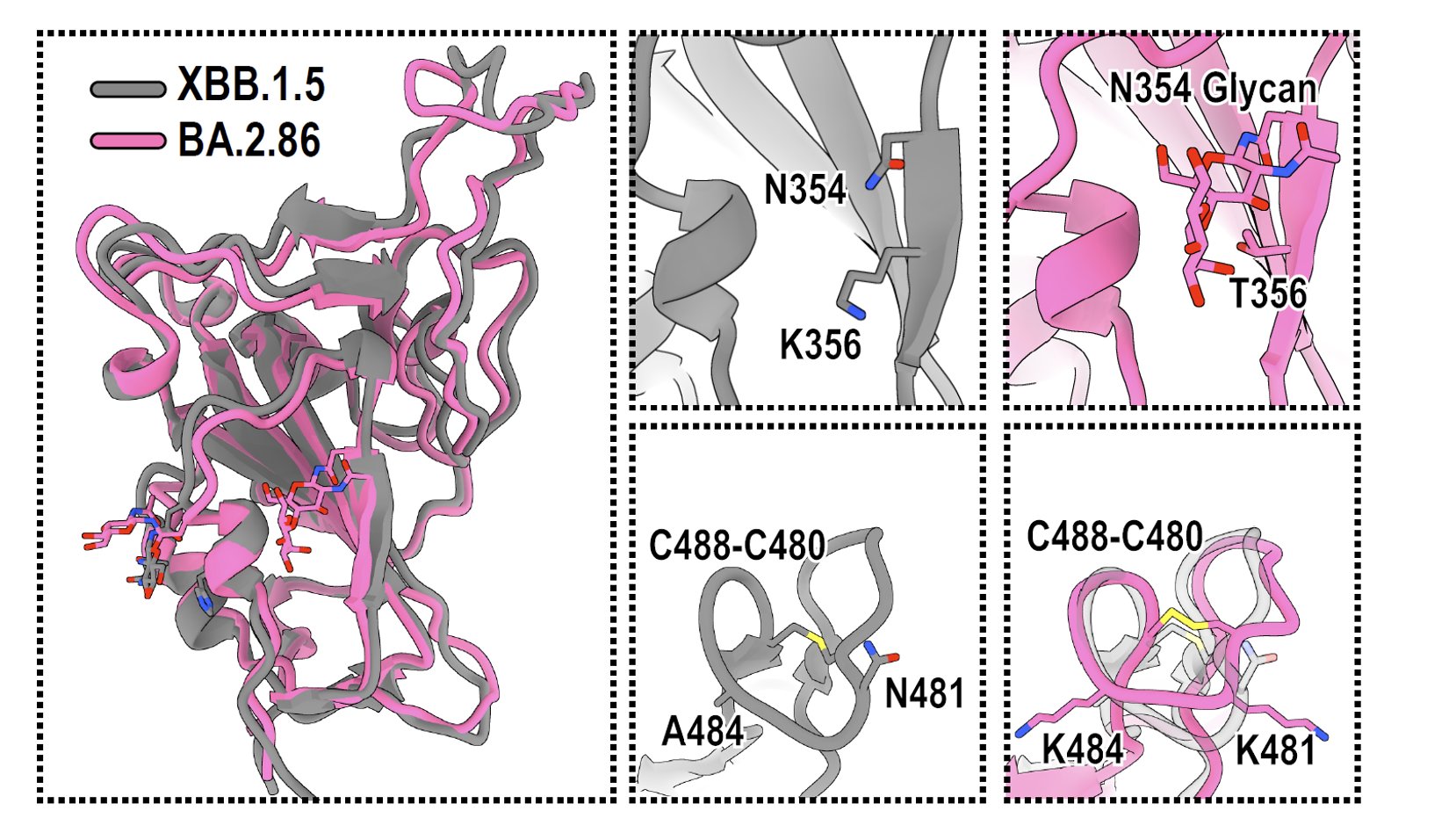
In sum, we showed that BA.2.86 exhibits high ACE2 binding, but probably still suffers from lower fusogenicity and infectivity (pseudovirus). The reason is likely due to BA.2.86's mutation on SD1-SD2 (E554K and P621S), which may cause BA.2.86's spike to favor "down" RBDs.
Various factors can determine a virus's transmission, including immune evasion, ACE2 binding, fusogenicity, etc. Since BA.1 can dominate given its reduced in vitro infectivity (proven in many studies), BA.2.86 can also prevail if it can break through immunity as good as BA.1 did.
So, can BA.2.86 break through current immunity as efficiently as BA.1 did before? Multiple BA.2.86 studies have suggested the opposite. In my opinion, the current form of BA.2.86 can probably beat XBB.1.5, maybe EG.5 as well, but not "FLip" variants like HK.3.
However, BA.2.86 is evolving (BA.2.86.1 already assigned), and that's why we need to closely monitor its evolution, to see whether BA.2.86 can gain mutations or reversions (especially on Spike) that could increase its transmissibility or immune evasion.

